2411
Automatic registration of MRI and transcranial ultrasound for the analysis of neurological disorders1PERFORM Centre, Concordia University, Montreal, QC, Canada, 2Department of Electrical and Computer Engineering, Concordia University, Montreal, QC, Canada, 3NeuroRx Research, Montreal, QC, Canada, 4Montreal Neurological Institute, McGill University, Montreal, QC, Canada, 5Department of Diagnostic and Interventional Neuroradiology, Montreal Neurological Hospital, Montreal, QC, Canada
Synopsis
Transcranial ultrasound (TCUS) can be used to diagnose and monitor a range of neurological conditions, such as Parkinson’s disease. However, reliable quantitative examination and multi-modal image analysis that involves TCUS require TCUS-MRI registration to guide the interpretation and measurement of the TCUS. We demonstrate that accurate rigid registration can be achieved through aligning gradient orientations of the 3D TCUS and an associated pseudo-TCUS constructed from the T1w MRI.
Purpose
Transcranial ultrasound (TCUS) is a special US technique that can image internal brain structures through acoustic windows of the human skull by operating at a low frequency (1∼5 MHz). With its advantages, such as low cost, high flexibility, and real-time imaging ability, TCUS has been popular for the diagnosis and monitoring of neurovascular conditions in the clinic. More recently, TCUS has shown promise to diagnose neurodegenerative conditions, such as Parkinson’s disease1 and Wilson’s disease2, by examining the abnormal image intensities in the image. However, the examination of TCUS images can be challenging because of lower image quality and difficult interpretation due to oblique imaging angles. To allow multi-modal image analysis and facilitate accurate assessment of the image features, TCUS and MR information must be aligned.Methods
Five healthy subjects (2 females, age=30±6 yo) were scanned with a 3D MPRAGE T1w MRI protocol (TE=3.4ms, TR=2.3s, TI=0.9s, flip angle = 9 degrees, resolution = 1x1x1 mm3) on a Siemens Tim Trio 3T MRI scanner. Series of spatially tracked 2D B-mode TCUS scans were obtained from temporal bone windows of each subject bilaterally. 3D TCUS volumes were reconstructed from the 2D images for both left and right sides. To allow for subject movement, head position was spatially tracked by an optical tracking system. An initial alignment between TCUS and MRI volumes was obtained by a rigid transformation through the alignment of corresponding facial landmarks of the MRI and the patient in real space. Then, a gradient-orientation-based registration technique3 was used to automatically register the 3D TCUS and a pseudo-TCUS constructed from the MRI data. First, cerebrospinal fluid (CSF) was segmented from each subject’s T1w MRI using a minimum distance classifier, and then the pseudo-TCUS Ipseudo was created through Ipseudo=1/(1+exp(0.2*D(Icsf))), where D(Icsf) is the distance transformation of CSF segmentation Icsf. The rigid transformation Treg was computed by registering the 3D TCUS to the pseudo-TCUS. To validate the registration performance, a ground truth transformation Tgt between 3D TCUS and the initially registered MR image was generated through manual alignment based on image feature correspondence by an expert. A 3D grid of regularly spaced (10 mm apart) landmarks P were defined within the brain region of the initially registered MRI of each subject, and the registration quality is evaluated as the root mean squared error (RMSE) between the point sets P and Treg* (Tgt)-1*PResults
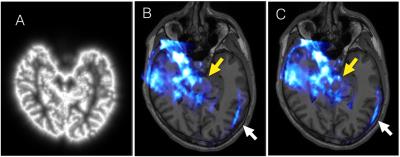
The pseudo-TCUS is demonstrated in Fig. 1A. TCUS volumes before and after registration that are overlaid on the T1w MRI (with the same view as Fig. 1A) are shown in Fig. 1B&C. The alignment of brainstem, ventricles and brain edges are visibly improved. Quantitatively, the RMSEs of the landmarks were evaluated at 1.89±0.14 mm and 1.86±0.47 mm, for the TUCS obtained from the left and right side respectively, and the RMSE for both sides was 1.88±0.32mm.Discussion
Initial alignment of TCUS and MRI through optical spatial tracking and facial landmark-based registration may not result in satisfactory TCUS-MRI alignment as shown in Fig 1B. In this case, additional registration is required to enable multi-modal image analysis. Although it is possible to achieve the alignment by directly using the T1w MRI, we have found that this approach failed in most of the cases by visual inspection. The pseudo-TCUS is created based on the observation that most hyperintense signals appear in TCUS correspond to the CSF. With the processed MRI, the registration performance is more robust against the variation of the TCUS image quality across different subjects for the chosen registration technique. TCUS can then be registered to any additional image modality through the T1w MRI indirectly.Conclusion
We have demonstrated that accurate intra-subject registration between TCUS and MRI can be achieved using an automatic gradient-orientation-based registration technique3 and pseudo-TCUS. The method can facilitate multi-modal image analysis of neurodegenerative diseases involving TCUS, opening new opportunities for efficient diagnostic techniques.Acknowledgements
No acknowledgement found.References
1. Behnke S. et al. Differentiation of Parkinson’s disease and atypical parkinsonian syndromes by transcranial ultrasound, J Neurol Neurosurg Psychiatry, 2005.
2. Svetel M. et al. Transcranial sonography in Wilson’s disease, Parkinsonism Relat Disord, 2012.
3. De Nigris D. et al., Fast rigid registration of pre-operative magnetic resonance images to intra-operative ultrasound for neurosurgery based on high confidence gradient orientations, IJCARS, 2013.