2292
Longitudinally consistent infant cortical surface atlases and parcellations from birth to 6 years of age1BRIC, UNC-Chapel Hill, Chapel Hill, NC, United States
Synopsis
For the first time, a longitudinally consistent infant cortical surface atlas with densely-sampled 11 time points (at 1, 3, 6, 9, 12, 18, 24, 36, 48, 60, and 72 months of age) is built for better exploring the dynamic and critical early brain development, based on 339 serial MRI scans from 50 healthy infants. The longitudinal consistency and unbiasedness are ensured by an advanced two-stage group-wise surface registration during the atlas construction. To equip parcellations for our atlases, both the FreeSurfer parcellation (for coarse parcellation) and HCP MMP parcellation (for fine-grained parcellation) are mapped onto our infant atlases.
Introduction
Longitudinal infant cortical surface atlases with dense sampling time-points and meaningful parcellations are of fundamental importance for understanding the dynamic and critical early brain development, but are still lacking. To fill this critical gap, in this study, for the first time, we construct longitudinal infant cortical surface atlases with totally 11 time points (at 1, 3, 6, 9, 12, 18, 24, 36, 48, 60, and 72 months of age) based on 339 serial MRI scans from 50 healthy infants. To ensure longitudinal consistency and unbiasedness, we adopt an advanced two-stage group-wise surface registration strategy. To equip parcellations with our infant atlases, we warp both the FreeSurfer parcellation1 with 35 regions in each hemisphere (for coarse parcellation) and HCP MMP parcellation2 with 180 regions in each hemisphere (for fine-grained parcellation) onto our atlases.Methods
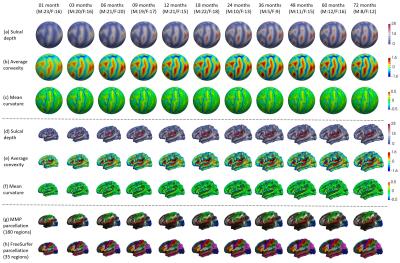
Totally 339 serial MRI scans from 50 healthy infants, each scheduled to be scanned at 1, 3, 6, 9, 12, 18, 24, 36, 48, 60, and 72 months of age, were used to construct our longitudinal infant cortical surface atlases. The subject number with gender information at each time point (with M indicating male, and F indicating female) is provided in Fig. 1. All infant MR images were processed by an established infant-specific computational pipeline3. Briefly, it included skull stripping, cerebellum removal, intensity inhomogeneity correction, tissue segmentation using a learning-based framework4, separation of left/right hemispheres, topology correction, inner and outer surface reconstruction, spherical mapping, and computation of cortical properties (e.g., sulcal depth, convexity, thickness, and curvature).
To build longitudinally consistent surface atlases, both intra-subject and inter-subject surface registrations were performed3. Specifically, to establish longitudinal cortical correspondences for each subject, first, the spherical surfaces of all time points for the same subject were registered together using an unbiased group-wise registration method5. Then, for each subject, a within-subject mean spherical surface was constructed by averaging corresponding cortical properties across all time points. Next, to establish inter-subject cortical correspondences, an unbiased group-wise registration was further performed to align the within-subject mean spherical surfaces of all different subjects. After that, for each age, an age-specific surface atlas consisting of the mean and variance of cortical properties across all infants at this age was constructed on the spherical surface, based on the inter-subject cortical correspondences defined above. Finally, a population-specific spherical surface atlas was obtained by computing the mean and variance of cortical properties across all within-subject mean surfaces.
To equip cortical parcellations with our infant atlases, the population-specific spherical surface atlas was aligned onto the FreeSurfer atlas. Then, for coarse parcellation, the FreeSurfer parcellation with 35 regions in each hemisphere1 was first propagated to our infant population-specific atlas and then further to our longitudinal infant surface atlases at all time points. For fine-grained parcellation, the HCP multi-modal parcellation (MMP) with 180 detailed regions in each hemisphere2 was first mapped to the FreeSurfer atlas using the HCP workbench6, and then propagated to our longitudinal infant surface atlases. The FreeSurfer parcellation based on sulcal-gyral patterns is well accepted by the community, while the MMP parcellation provides more insights into the cortex from both anatomical and functional points of view.
Results
Fig. 1 shows the constructed longitudinal infant surface atlases for the left hemisphere at each time point. Rows (a) – (c) illustrate the cortical properties on the standard sphere; Rows (d) – (e) illustrate the corresponding cortical properties on the age-specific mean cortical surface; Rows (f) and (g) illustrate the MMP parcellation and the FreeSurfer parcellation at each time point. From this figure, we can find that, the infant brain forms the major cortical folding patterns at birth, which are largely preserved during postnatal development. Also we can see that the cortical properties (i.e., sulcal depth, convexity, curvature and surface area) have rapid changes as the age increases. For example, we show the dramatic development of sulcal depth from 1 month to 24 months in Fig. 1 (a) and (d). After 24 months, many cortical properties have only subtle changes. Meanwhile, our infant surface atlases are longitudinally consistent in terms of cortical properties and parcellations.Conclusion
We have proposed a new method to construct longitudinally-consistent infant cortical surface atlases by using two-stage groupwise registration. Leveraging this method and our unique longitudinal infant MRI dataset, we have constructed the first longitudinal infant cortical surface atlases with densely-sampled 11 time points at 1, 3, 6, 9, 12, 18, 24, 36, 48, 60, and 72 months of age, with both FreeSurfer and MMP parcellations. We will release our infant surface atlases and parcellations to the public.Acknowledgements
This work was supported in part by NIH grants (MH100217, MH108914 and MH107815).References
[1]. Desikan, R.S., F. Ségonne, B. Fischl, et al., An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage, 2006;31(3):968-980.
[2]. Glasser, M.F., T.S. Coalson, E.C. Robinson, et al., A multi-modal parcellation of human cerebral cortex. Nature, 2016;536(7615):171-178.
[3]. Li, G., L. Wang, F. Shi, et al., Construction of 4D high-definition cortical surface atlases of infants: Methods and applications. Medical image analysis, 2015;25(1):22-36.
[4]. Wang, L., Y. Gao, F. Shi, et al., LINKS: Learning-based multi-source IntegratioN frameworK for Segmentation of infant brain images. NeuroImage, 2015;108:160-172.
[5]. Yeo, B.T., M.R. Sabuncu, T. Vercauteren, et al., Spherical demons: fast diffeomorphic landmark-free surface registration. IEEE transactions on medical imaging, 2010;29(3):650-668.
[6]. Van Essen, D.C., S.M. Smith, D.M. Barch, et al., The WU-Minn human connectome project: an overview. Neuroimage, 2013;80:62-79.