2281
A pilot study of lateral ventricle volume from in utero foetal brain magnetic resonance imaging (MRI)1State Key Laboratory of Bioelectronics, Southeast University, Nanjing, People's Republic of China, 2School of Biological Science and Medical Engineering, Southeast University, Nanjing, People's Republic of China, 3The First School of Clinical Medicine, Nanjing Medical University, Nanjing, People's Republic of China, 4Department of Radiology, Drum Tower Hospital of Nanjing University Medical School, Nanjing, People's Republic of China, 5Southeast University - Jiangsu Institute of Biomaterials and Medical Devices, Nanjing, People's Republic of China, 6Institute of Cancer and Genomic Science, University of Birmingham, United Kingdom
Synopsis
Neuroimaging for foetus brain is a challenging problem in which there are several issues to be solved. We proposed a general solution and finally reconstruct the lateral ventricle volume that is of great significance for clinical study. Firstly, slices were realigned to correct for the motion between acquisition of individual slices including transposition and rotation. Secondly, slices with motion artefacts were excluded and inconsistencies in intensity patterns resulting from the motion were estimated and corrected for. Thirdly, the structure of lateral ventricle was segmented via adaptive segmentation. Finally, the volume was reconstructed from irregularly sampled data.
Introduction
The foetal brain lateral ventricle structure shows great significance for antenatal diagnosis,1 and the magnetic resonance imaging (MRI) is adopted due to its relatively higher resolution compared with computer tomography and ultra sound.2 However, image reconstruction from MRI data for foetal brain volume becomes challenging caused by freely moving foetal head during data acquisition. Ultra-fast multi-slices MR sequences, such as Half Fourier Acquisition Single-Shot Turbo Spin Echo (HASTE) or Single-Shot Fast Spin Echo (ssFSE),3 were involved in clinical MRI examination in order to avoid as much as possible motion. Several image stacks that are often acquired in different orthogonal views are able to provide in-vivo 3D foetal anatomy structure in a short time that the influence of motion artefact can be relatively decreased. Super resolution techniques are introduced to combine the three low-resolution (LR) image stack in order to realise a high-resolution (HR) image stack. However, motion artefacts endure and are likely to generate inter-slice noise. Several image processing4 and sequence improvement5 approaches then become essential to realise high resolution imaging, including slice realignment to filter the motion, segmentation of the lateral ventricle structure and visualisation via curve filtering. Some efficient algorithms are proposed to improve the imaging quality, such as registration,6 super resolution,7-9 total variation,10 compressed sensing11 and so on, yet an automated processing framework is still in need helping doctors to visualise and evaluate the volume.Methods
In this paper, we acquired an amount of MR image datasets using Philips Multiva (PHILIPS-M36JUBL) 1.5 Tesla two-dimensional balanced turbo field echo (BTFE) sequence, with the gestation week from 26 to 37, slice thickness of 6 to 7mm, flip angle of 90 degree, imaging frequency of 63.88, slice spacing of 1mm, echo time of 2.783ms, slice amount of 60 to 80, repetition time of 5.566ms, field of view of 250*250*66mm, reconstruction voxel size of 0.391mm, reconstruction matrix of 640mm, acquisition time of 80 to 100s, pixel spacing of 0.390625\0.390625. We model the imaging procedure as a linear model, in which the motion, blur, downsampling and bias operators were combined as the noise operator from the original high-resolution (HR) image stack $$$x$$$ to the observed low-resolution (LR) image stack $$$x_k^{\text{LR}}$$$. The data acquisition procedure can be denoted as
$$x_k^{\text{LR}}=L_kG_kB_kM_kx+n_k~\sim H_kx+n_k$$
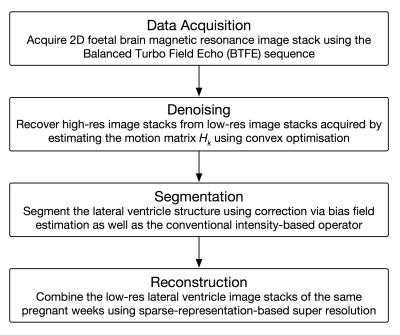
Based on this model, we process the image datasets as following, and the flowchart of our approach is given by Figure 1. Firstly, the slices in the image stacks were realigned using convex optimisation based on total variation (TV) semi-norm,12 by which the motion artefact were decreased relatively.
$$\min_{x\in X} ||x||_{\text{TV}}+\frac{\lambda}{2}\sum_{k=1}^K ||H_kx-x_k^{\text{LR}}||^2\text{ s.t. }x\geq 0$$
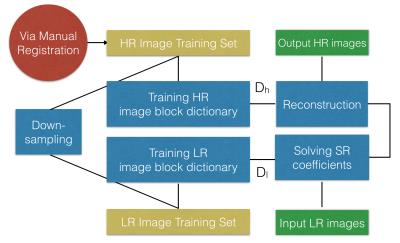
Secondly, the lateral ventricle structure was segmented using adaptive segmentation,13 in which the bias field was estimated using Bayesian method for conventional intensity-based segmentation.14 Thirdly, the image stacks of the same gestation week were combined using sparse representation15 and registration via Elastix16, by which the super resolution reconstruction for lateral ventricle volume was realised. The procedure of super resolution reconstruction is given by Figure 2.
$$\min ||\alpha||_0\mbox{ s.t. }||FD_l\alpha-Fy||^2_2\leq \epsilon$$
Finally, the reconstructed volume was interpolated and smoothed using Bézier plane.17 Our approach implemented the foetal brain volume development shown by image reconstruction as well as several filtering methods aiming to improve the accuracy.
Results
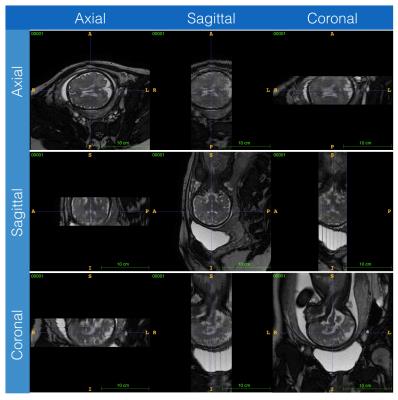
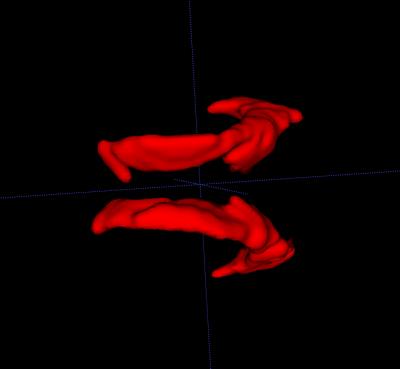
Visualised volume of the lateral ventricle is reconstructed and the statistical result of the volume development is achieved varying among gestation weeks, as well as the difference between the abnormal and normal subjects also compared. The data acquired in the three orthogonal directions and the reconstructed volume of lateral ventricle are given by Figure 3 and 4, respectively. The volume helps to show the development character of the foetal brain.Conclusion
Our results shows the development of the foetal brain volume with gestation variation as well as its difference between abnormal and normal ones, which can be used as reference for antenatal diagnosis. Compared with the previous work, our results provide a distinct model for foetal brain observation with an ameliorated signal-to-noise ratio (SNR). In the future, more registration and segmentation methods will be studied and more smoothing curve fitting approach will be observed. It is expected to associate the proposed algorithms as an automated image post-processing and visualisation framework for clinical study. On the other hand, more datasets will be acquired to enhance the accuracy and reliability of our modelling.Acknowledgements
The authors thank department of radiology, the affiliated Drum Tower Hospital of Nanjing University Medical School for the MRI data acquisition, the Shanghai United Imaging Inc. and Southeast University for the maintenance of the MRI system.References
1. Huang H, Xue R, Zhang J, Ren T, Richards LJ, Yarowsky P, Miller MI, Mori S. Anatomical Characterization of Human Fetal Brain Development with Diffusion Tensor Magnetic Resonance Imaging. Journal of Neuroscience 2009;29(13):4263–4273.
2. Glenn OA. MR imaging of the fetal brain. Pediatr Radiol 2009;40(1):68–81.
3. Levine, D., Hatabu, H., Gaa, J., Atkinson, M., Edelman, R. Fetal anatomy revealed with fast mr sequences. American Journal of Roentgenology 1996;167(4):905–908.
4. Malamateniou C, Malik SJ, Counsell SJ, et al. Motion-Compensation Techniques in Neonatal and Fetal MR Imaging. American Journal of Neuroradiology 2013;34(6):1124–1136.
5. Griffiths PD, Jarvis D, McQuillan H, Williams F, Paley M, Armitage P. MRI of the foetal brain using a rapid 3D steady-state sequence. BJR 2013;86(1030):20130168–8.
6. Francois R, Orit G, Bistra I, Claudia R, Daniel V, James B, Colin S, Registration-based approach for reconstruction of high-resolution in utero foetal MR brain images. Academic Radiology. 2006;13(9):1072-1081.
7. Rousseau F, Kim K, Studholme C, Koob M, Dietemann JL. On super-resolution for fetal brain MRI. Med Image Comput Comput Assist Interv 2010;13(2):355–362.
8. Jiang S, Xue H, Glover A, Rutherford M, Rueckert D, Hajnal JV. MRI of Moving Subjects Using Multislice Snapshot Images With Volume Reconstruction (SVR): Application to Fetal, Neonatal, and Adult Brain Studies. IEEE Trans. Med. Imaging 2007;26(7):967–980.
9. Gholipour A, Estroff JA, Warfield SK. Robust Super-Resolution Volume Reconstruction From Slice Acquisitions: Application to Fetal Brain MRI. IEEE Trans. Med. Imaging 2010;29(10):1739–1758.
10. Tourbier S, Bresson X, Hagmann P, Thiran J-P, Meuli R, Cuadra MB. Efficient total variation algorithm for fetal brain MRI reconstruction. Med Image Comput Comput Assist Interv 2014;17(2):252–259.
11. Ning L, Setsompop K, Michailovich O, Makris N, Shenton ME, Westin C-F, Rathi Y. A joint compressed-sensing and super-resolution approach for very high-resolution diffusion imaging. NeuroImage 2016;125(15):386–400.
12. Chambolle A, Pock T. A First-Order Primal-Dual Algorithm for Convex Problems with Applications to Imaging. J Math Imaging Vis 2010;40(1):120–145.
13. Wells WM, Grimson WEL, Kikinis R, Jolesz FA. Adaptive segmentation of MRI data. IEEE Trans. Med. Imaging 1996;15(4):429–442.
14. Kuklisova-Murgasova M, Quaghebeur G, Rutherford MA, Hajnal JV, Schnabel JA. Reconstruction of fetal brain MRI with intensity matching and complete outlier removal. Medical Image Analysis 2012;16(8):1550–1564.
15. Yang J, Wright J, Huang TS, Ma Y. Image Super-Resolution Via Sparse Representation. IEEE Transactions on Image Processing 2010;19(11):2861–2873.
16. Hahmann S, Bonneau GP. Polynomial surfaces interpolating arbitrary triangulations. IEEE Trans. Visual. Comput. Graphics 2003;9(1):99–109.
Figures