2279
Fast and Robust Detection of Fetal Brain in MRI using Transfer Learning based FCN1Shenzhen Research Institute, The Chinese University of Hong Kong, Shenzhen, People's Republic of China, 2Department of Imaging and Interventional Radiology, The Chinese University of Hong Kong, Hong Kong, Hong Kong, 3Shenzhen Research Institute,The Chinese University of Hong Kong, Shenzhen, People's Republic of China, 4Department of Radiology, Nanjing Drum Tower Hospital,The Affiliated Hospital of Nanjing University Medical School, Nanjing, People's Republic of China, 5Philips Healthcare, Hong Kong, 6Chow Yuk Ho Technology Center for Innovative Medicine, Hong Kong, 7Therese Pei Fong Chow Research Centre for Prevention of Dementia, 8Department of Medicine and Therapeutics, The Chinese University of Hong Kong, Hong Kong
Synopsis
In this work, we proposed a transfer learning based FCN method which can automatically detect the fetal brain in MRI. We used the off-the-shelf model weights trained on nature images to initialize a fully connected network (FCN), and then fine-tuned the model on the fetal MRIs. We tested our method on two datasets with different MRI sequences, and the results demonstrated that the proposed method is automatic, fast and robust for detection of fetal brain in MRI.
Purpose
MRI is increasingly used in prenatal diagnosis in recent years. The high resolution characteristic of MRI makes it possible of examining the development status of fetal brain. Currently, the clinicians have to manually localize and segment the fetal brain in MRI for the brain reconstruction and further analysis. The manual operation is time-consuming, so it is urgent to develop an automatic method to perform this task. But the diverse MR images which are from different sequence, different machines and different centers make the automatic detection challenging. With the machine learning methods emerging in computer vision and gradually applied in medical images, it is practicable to automatically detect and segment the organs or regions of interest in medical images to avoid manual operation and potentially inconsistent annotations among radiologists. The objective of this paper is to propose an automatic, fast and robust method for detection of fetal brain in MRI.Methods
A fully connected network (FCN)[1] which can directly learn the visual features from the raw images without any preprocessing was used to produce the fetal brain probability map on each MRI slice in the axial plane. Then the probability map was threshold by 0.5 to get the binary brain mask. Finally, to clear up the small false positive regions, only the biggest connected component region was remained to get the bounding box in the final detection results. FCN was composed of multiple convolutional layers and pooling layers with millions of parameters, which made it reach full abilities only under a large number of annotated training data. However, in the context of fetal MRI, the number of annotated images was insufficient to train this model. So the transfer learning[2] method was applied by using the off-the-shelf model weights[3] trained on a huge number of labelled nature images to initialize our FCN and then fine-tuning the model on the fetal MRIs. The transfer learning not only made the FCN working on small dataset possible, but also the pre-trained weights containing abundant low-level visual features learned from the natural images made the FCN more robust on MRI with other sequences.Results
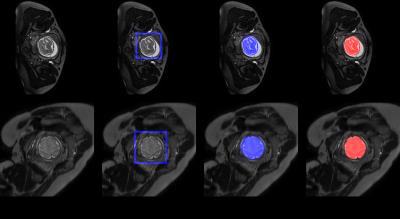
88 T2-weighted MRIs of fetuses among 20 to 30 gestational week were collected using a Philips 1.5T machine with B-FFE sequence (TR=1~6 ms, TE = 1~6 ms, flip angle = 90°, slice thickness = 7 mm, in-plane matrix size = 288×288, FOV = 379×253×103 mm, slice thickness/gap = 2.5/1.5 mm with interleaved acquisition, slice number = 22) from Nanjing Drum Tower Hospital, China, and the locations of the brains were annotated the by the experienced radiologists. The images were splitted into training, validation and testing sets which contained 44, 11 and 33 images, respectively. The validation dataset was used to select the model with the best generalization ability. The detection accuracy was 100% and the average DICE was 87.8% on the testing dataset. And it cost only 2 seconds to detect the whole fetal brain on one subject. Besides, to test the robustness of the model, the model was also applied on the other 10 T2-weighted MRIs with different sequence which were acquired using a Philips Multiva 1.5T machine with SSFSE sequence (TR=4500ms, TE=91ms, flip angle=90°, in-plane matrix size=400×400, FOV=300×300mm, slice thickness/gap=3/0mm with interleaved acquisition, slice number=30). The average detection accuracy on these MRIs was also 100%. The examples of our results on both two MRI sequence are in the Fig. 1.Discussion
The results from our experiments demonstrate that transfer learning based FCN is fast, robust and high accurate on detection of fetal brain in MRI with only few training data. The high performance of our method will make it as a useful tool in both areas of clinical usage and research. In the future, we will conduct more experiments on other fetal MRI sequences.Conclusion
In this work, we presented a transfer learning based FCN method which can automatically detect the fetal brain in MRI. This method overcame the problem of limited annotated images which always hinders applying machine learning methods in medical images, and it can be applied to other medical image tasks.Acknowledgements
No acknowledgement found.References
[1]Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation[C]//Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015: 3431-3440.
[2] Shin H C, Roth H R, Gao M, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning[J]. IEEE transactions on medical imaging, 2016, 35(5): 1285-1298.
[3] Chen L C, Papandreou G, Kokkinos I, et al. Semantic image segmentation with deep convolutional nets and fully connected crfs[J]. arXiv preprint arXiv:1412.7062, 2014.
Figures