2236
Aberrant striatal anatomy and gray matter connectivity networks in mice lacking autism-associated gene CNTNAP2.1Functional Neuroimaging Laboratory, Center for Neuroscience and Cognitive Systems, Istituto Italiano di Tecnologia, Rovereto, Italy, 2CIMeC, Center for Mind/Brain Sciences, University of Trento, Rovereto, Italy
Synopsis
Mice lacking CNTNAP2 exhibit robust autism-like behavioral traits, including stereotyped behaviors and excessive self-grooming. By using high resolution morpho-anatomical imaging in CNTNAP2 mutant mice, we identified marked volumetric alterations subcortical substrates implicated in ASD motor stereotypy, with a prominent involvement of the dorsal striatum. Importantly, we also show that in mutant mice, striatal but not cortical regions, exhibit dramatically expanded gray matter network extension, encompassing aberrant trophic interaction between limbic, subcortical and prefrontal regions. The observed striatal volumetric and gray matter network abnormalities serve as a plausible morpho-anatomical substrate for some of the stereotypy exhibited by CNTNAP2 mutant mice.
INTRODUCTION
CNTNAP2 is an autism-associated gene1-4 that encodes CASPR2, a trans-membrane cell-adhesion protein involved in the clustering of K+ channels5,6. CNTNAP2 deficient mice exhibit decrease in dendritic arborization and spine development7, as well as autistic-like motor stereotypy and hyperactivity8, and reduced prefrontal connectivity9. The broad spectrum of phenotypic alteration observed in carriers of homozygous CNTNAP2 mutations10 suggests the involvement of distributed brain substrates in the phenotypic expression of this condition. However, attempts to identify the brainwide substrates affected by CNTANP2 via imaging of human carriers of CNTNAP2 polymorphisms, have produced inconsistent result, possibly reflecting clinical heterogeneity in the mutations mapped11,12. As a result of this, the brainwide anatomical substrates affected by homozygous loss of CNTNAP2, a condition with high autism penetrance, remain undetermined. To probe a mechanistic involvement of CNTNAP2 in shaping gray matter (GM) volume and trophic dynamics occurring between connected brain regions - known as structural covariance MRI (scMRI) networks - we used high resolution structural MRI in Cntnap2-/- and control mice13,14. The approach revealed aberrant striatal anatomy and GM connectivity networks in CNTNAP2 mice, which serve as a plausible morpho-anatomical substrate for some of the stereotypy produced by this mutation.METHODS
High-resolution morpho-anatomical T2-weighted MR imaging of ex-vivo Cntnap+/+ (n=10) and Cntnap-/- (n=14) mouse brains was performed at 7 Tesla, using a FLASH sequence with an isotropic voxel size of 70 µm s recently described14. Inter-group differences in local volumes were mapped with VBM and volumetric anatomical labelling13. To confirm regional location of morphometric alterations in the striatum, a vertex-wise shape analysis based on spherical harmonics was employed15. We also used seed-based mapping (t>3.5, pc=0.01) to probe growth dynamics between brain regions (structural covariance network mapping - scMRI). scMRI networks between eighteen representative neuroanatomical volumes were clustered using the k-means method14.RESULTS
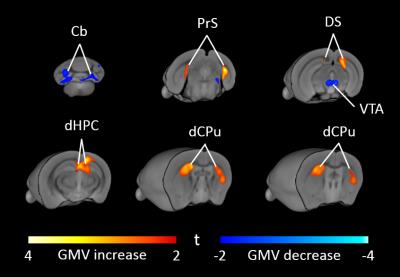
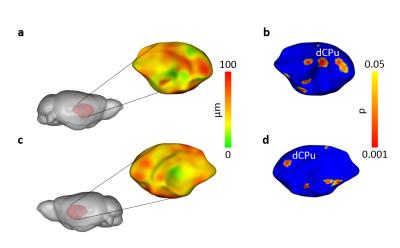
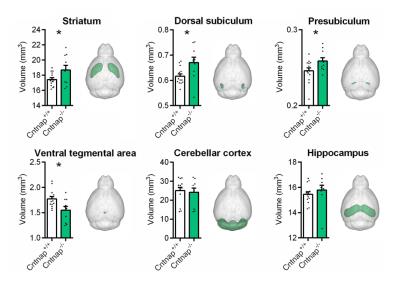
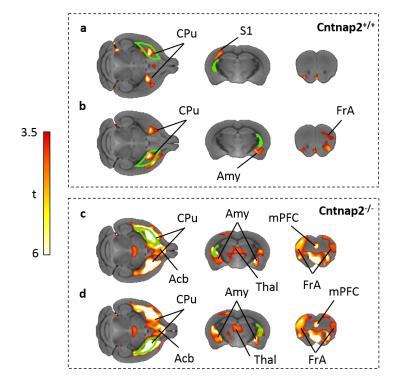
High-resolution VBM mapping revealed focal bilateral areas of increased GM volume in dorsal striatum and hippocampal formation, as well as a decrease GM volume in the cerebellum and ventral tegmental area in Cntnap2-/- mice compared to wild-type littermates (t> |2|, pc<0.01, Fig.1). Importantly, striatal shape analysis revealed the presence of aberrant striatal enlargement in Cntnap2-/- mice, with an outward bilateral displacements in the dorsomedial surface of this structure, a key site for habitual and repetitive behaviour in human and rodents16,17 (Fig.2). Analogous effects were observed using automated anatomical labelling (Fig.3).
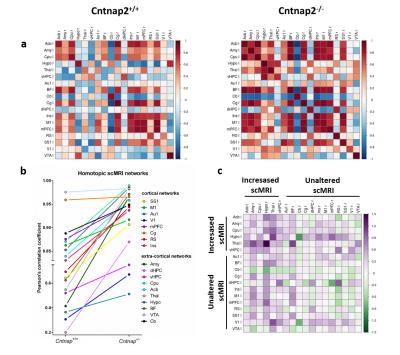
GM connectivity network matrices for Cntnap2-/- and control mice are shown in Fig. 4. scMRI mapping of homotopic GM connectivity revealed increased inter-hemispheric connectivity across all the regions examined in Cntnap2-/- mice with respect to control littermates (Fig.4). However, when regions were clustered based on their heterotopic connectivity, two distinct sets of regions were identified. Of particular relevance was a set of subcortical regions including the striatum, amygdala, thalamus, which appeared to exhibit large heterotopic connectivity than all the rest of cortical regions. This effect was apparent when scMRI network of the striatum was mapped using seed-based approach (Fig. 5), revealing a dramatic reorganization of the striatal GM covariance in mutants with respect to control mice, with an aberrant involvement of subcortical and prefrontal substrates that was not present in control mice. This results suggest that the striatal hypertrophism observed in CNTNAP2 mutants may affect the coordinated growth of a set of connected regions, resulting in brainwide aberrancies in GM networks organization.
CONCLUSIONS
Here, we show that homozygous mice lacking CNTNAP2 exhibit macroscale volumetric alterations in the striatum and other subcortical regions previously associated to ASD. Our data also provide a mechanistic link between CNTNAP2 deficiency and scMRI dynamics, supporting the view that hyperactivity and motor stereotypy exhibited by these mice could be generated by network-level interactions between striatum and neighbouring subcortical regions, which serve as a plausible morpho-anatomical substrate for some of the stereotypy produced by this mutation. We expect this observation to spur analogous multivariate GM analyses in human carriers of CNTANP2 mutations.Acknowledgements
The study was funded by a grant from the Simons Foundation (SFARI 314688, A.G.)References
1.Arking, D.E., et al., A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. The American Journal of Human Genetics, 2008. 82(1): p. 160-164.
2.Anney, R., et al., Individual common variants exert weak effects on the risk for autism spectrum disorders. Human molecular genetics, 2012. 21(21): p. 4781-4792.
3.Bakkaloglu, B., et al., Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. The American Journal of Human Genetics, 2008. 82(1): p. 165-173.
4.Alarcón, M., et al., Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. The American Journal of Human Genetics, 2008. 82(1): p. 150-159.
5.Poliak, S., et al., Caspr2, a new member of the neurexin superfamily, is localized at the juxtaparanodes of myelinated axons and associates with K+ channels. Neuron, 1999. 24(4): p. 1037-1047.
6.Poliak, S., et al., Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. The Journal of cell biology, 2003. 162(6): p. 1149-1160.
7.Anderson, G.R., et al., Candidate autism gene screen identifies critical role for cell-adhesion molecule CASPR2 in dendritic arborization and spine development. Proceedings of the National Academy of Sciences, 2012. 109(44): p. 18120-18125.
8.Peñagarikano, O., et al., Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell, 2011. 147(1): p. 235-246.
9.Liska, A., et al., Homozygous loss of autism-risk gene CNTNAP2 results in reduced local and long-range prefrontal functional connectivity. bioRxiv, 2016.
10.Rodenas-Cuadrado, P., J. Ho, and S.C. Vernes, Shining a light on CNTNAP2: complex functions to complex disorders. European Journal of Human Genetics, 2014. 22(2): p. 171-178.
11.Tan, G.C., et al., Normal variation in fronto-occipital circuitry and cerebellar structure with an autism-associated polymorphism of CNTNAP2. Neuroimage, 2010. 53(3): p. 1030-1042.
12.Uddén, J., et al., A common variant of the CNTNAP2 gene is associated with structural variation in the left superior occipital gyrus. Brain and language, 2016.
13.Pagani, M., et al., Semi-automated registration-based anatomical labelling, voxel based morphometry and cortical thickness mapping of the mouse brain. Journal of Neuroscience Methods, 2016. 267: p. 62-73.
14.Pagani, M., A. Bifone, and A. Gozzi, Structural covariance networks in the mouse brain. Neuroimage, 2016.
15.Styner, M., et al., Framework for the statistical shape analysis of brain structures using SPHARM-PDM. The insight journal, 2006(1071): p. 242.
16.Hollander, E., et al., Striatal volume on magnetic resonance imaging and repetitive behaviors in autism. Biol Psychiatry, 2005. 58(3): p. 226-232.
17.Langen, M., et al., The neurobiology of repetitive behavior: of mice…. Neuroscience & Biobehavioral Reviews, 2011. 35(3): p. 345-355.
18.Pagani, M., A. Bifone, and A. Gozzi, Structural covariance networks in the mouse brain. NeuroImage, 2016. 129: p. 55-63.
Figures