2220
Radiographic Atlas of Tumor Growth Pattern in GlioblastomaMorteza Esmaeili1, Anne Line Stensjøen1, Erik Magnus Berntsen1, Ole Solheim1,2,3, and Ingerid Reinertsen3,4
1Department of Circulation and Medical Imaging, Norwegian University of Science and Technology, NTNU, Trondheim, Norway, 2Department of Neurosurgery, St. Olav’s University Hospital, Trondheim, Norway, 3Norwegian National Advisory unit for Ultrasound and Image Guided Therapy, St. Olav’s University Hospital, Trondheim, Norway, 4Department of Medical Technology, SINTEF, Trondheim, Norway
Synopsis
In this study we assess the growth pattern of one of the most aggressive brain tumor in adults. Surgery is a standard of care for these patients, and understanding the prominent growth direction of tumor lesions can eventually benefit surgical planning. Contrast-enhanced MR data at two time-points of diagnosis and pre-operation were analyzed to derive mean 3D vector field demonstrating the growth directions. A DTI white matter (WM) atlas was used to investigate the degree of agreement and alignment of the generated vector field towards white matter fibers.
INTRODUCTION
Derived from glial cells, glioblastoma multiforme (GBM) is an extremely aggressive malignant tumor in adults. Generating MR-derived growth pattern models for GBM have been of an attractive approach in neuro-oncology, suggesting a distinct pattern of lesion spread with tendency in growing along the white fiber direction for the invasive component (1,2). Here we report preliminary results from a retrospective analysis, designed to provide a brain atlas predicting the dominant directions of tumor lesion expansion/shrinkage prior to surgery.METHODS
Forty three adult glioblastoma patients were examined at two time-points; diagnostic and pre-operation, with minimum 14 days between acquisitions with contrast-enhanced anatomical MRI (ce-T1). The study was approved by the regional ethics committee and adhered to the Helsinki Declaration. The tumor volumes were delineated by two radiology experts. The image analysis software BrainVoyager QX (Version 2.3, Brain Innovation B.V., Maastricht, The Netherlands) was used for semi-automatic tumor segmentation. The widely accepted Advanced Normalization Tools (ANTs) script packages (3) and customized MATLAB scripts were used for registration of MR images. The 3D data analysis pipeline (Fig. 1) was started with a few pre-processing steps including up-sampling for identical voxel number/size, N4 bias field correction (from ANTs package) and skull stripping using Brain Extraction Tool (BET) (4). Using segmented tumor masks, fine non-linear registration was performed to generate deformation fields of tumor lesions delineated from T1-diagnostic and T1-pre-operation images. Using non-linear transformation, all solutions were transformed into the MNI template space and compared with a DTI WM atlas (5). Angles between the vector fields of DTI WM and tumor growth directions were calculated (within sub-regions with prominent lesion overlaps) to estimate an angle map of lesion growth to that of white matter tracts.RESULTS
To compute the degree of alignment between
tumor growth vector fields and white matter tracts, an angle map was calculated.
The results of the generated maps were color-coded to visualize the alignment
agreements between two dataset, as yellow color indicates the maximum alignment
(θ<20º or θ>160º, Fig. 1). In all sub-regions, tumor displacement showed a
reproducible tendency in moving along the white matter tracts (Fig. 1), as evidenced
by voxel-wise parallel alignments of the mean vector fields towards the tensor field
of the WM atlas. CONCLUSION
The resulting GBM atlas of deformation vector field provides information on tumor growth pattern prior to surgery. We mapped the delineated growth pattern on DTI WM atlas, investigating the hypothesis that tumor cells tend to track along the white matter. Our findings represent a first step in investigating the hypothesis that tumor cells tend to track along the white matter.Acknowledgements
References
1. Stensjoen AL, Solheim O, Kvistad KA, Haberg AK, Salvesen O, Berntsen EM. Growth dynamics of untreated glioblastomas in vivo. Neuro Oncol 2015;17(10):1402-1411. 2. Bondiau PY, Clatz O, Sermesant M, Marcy PY, Delingette H, Frenay M, Ayache N. Biocomputing: numerical simulation of glioblastoma growth using diffusion tensor imaging. Phys Med Biol 2008;53(4):879-893. 3. Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 2010;29(6):1310-1320. 4. Muschelli J, Ullman NL, Mould WA, Vespa P, Hanley DF, Crainiceanu CM. Validated automatic brain extraction of head CT images. Neuroimage 2015;114:379-385. 5. Zhang H, Awate SP, Das SR, Woo JH, Melhem ER, Gee JC, Yushkevich PA. A tract-specific framework for white matter morphometry combining macroscopic and microscopic tract features. Med Image Anal 2010;14(5):666-673.Figures
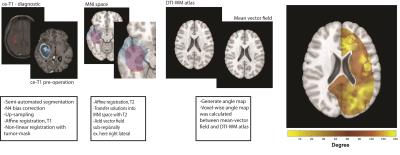
Figure 1. Schematic of the vector field
analysis pipeline. The intensity and direction of arrows demonstrate the distance
and direction of tumor movement respectively. The degree of alignment between the
3-dimensional mean vector field and a DTI-WM atlas was calculated and demonstrated as an angle map. The angle map demonstrates the tendency of tumor growth along WM fibers
(increased tendency from red to yellow).