2181
Learning-based Segmentation for Monkey Brain MRI1Department of Information Engineering, Liuzhou City Vocational College, Liuzhou, People's Republic of China, 2Department of Radiology and BRIC, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
Synopsis
Accurate segmentation of monkey brain MRI is of great importance in studying the brain development, pathogenesis and progression of neurological diseases. However, it is challenging for automatic segmentation due to noise, low
Introduction
Monkeys are close to humans physiologically and behaviorally, justifying their wide usage in studies that cannot be done on humans. Accurate segmentation of monkey brain MRI is of great importance in studying the brain development, pathogenesis and progression of neurological diseases. However, it is challenging for automatic segmentation due to noise, low contrast and partial volume effect. Existing tools were mainly fine-tuned to human brain MRI1,2,3, which is quite different from monkey brain MRI. Consequently, the accuracy is limited when applied to monkeys. Most previous works employed atlas-guided segmentation for monkey images, which are highly dependent on the accuracy of registration. In this abstract, we propose a machine learning-based framework for segmentation of monkey brain MRI into the skull, cerebellum, white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF) of the cerebrum.Methods
MRI data of 15 monkey brains is used in this study. The ages of these monkeys range from 12 months to 61 months. T1-weighted MR images were acquired by a Philips 3T whole-body MR system, with spatial resolution of 0.375×0.375×0.5 mm3. These 15 subjects were manually labeled by an experienced expert, and then used as the ground truth for training and evaluation of our classifiers. Specifically, inspired by Wang et al.’s work4, we use random forest to train a sequence of classifiers. Firstly, we train the first-layer classifiers by extracting appearance features from intensity images. The estimated probabilities maps of different regions are further employed to extract informative context features4 for training the next-layer classifier, together with the appearance features from the intensity images. Finally, a sequence of classifiers can be trained for accurate classification. During the training of each layer classifier, we randomly select 20000 samples for each label and generate 10000 3D Haar-like features for each sample. The setting of random forest includes the number of decision trees (i.e., 20), maximal depth (i.e., 50), and minimal samples in a leaf node (i.e., 8).Results
Fig. 1 shows the intermediate results on a typical subject. It can be seen that the probability maps are gradually improved with iterations and become more and more accurate.
Since the skull is one of classified labels, our proposed method can automatically perform skull stripping (Fig. 1). To demonstrate the advantage, we make comparisons with the popular skull-stripper, i.e., BET1, as shown in Fig. 2. It can be seen that our proposed method can obtain more accurate brain extraction, compared with the results by BET.
In the following, we further make comparisons with a conventional method, in which skull stripping, cerebellum removal and tissue segmentation are typically performed in a sequential way, instead of a simultaneous way. The 3-D renderings of WM by the conventional method4 and proposed method are shown in Fig. 3. It is clearly shown that some parts of skull are incorrectly classified as WM by the conventional method. Finally, Dice ratios are shown in Fig. 4, which indicate that our proposed method performs better than the conventional method.
Conclusions
A learning-based method has been presented for segmentation of monkey brain MRI. Comparison results demonstrated the advantage of our proposed method in terms of segmentation accuracy.Acknowledgements
This work was supported in part by National Institutes of Health grants (MH100217, MH108914 and MH107815).References
1. Smith, Stephen M., Fast robust automated brain extraction, Human Brain Mapping 2002, 17(3):143-155.
2. Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004, 23:S208-219.
3. J. Ashburner, K.J. Friston Voxel-based morphometry: the methods, NeuroImage, 2000,11: 805–821
4. Wang, Li, Yaozong Gao, Feng Shi, Gang Li, John H. Gilmore, Weili Lin, and Dinggang Shen. "LINKS: Learning-based multi-source Integration framework for Segmentation of infant brain images." NeuroImage. 2015, 108: 160-172.
Figures

Fig. 1. The probability maps produced by our proposed method.
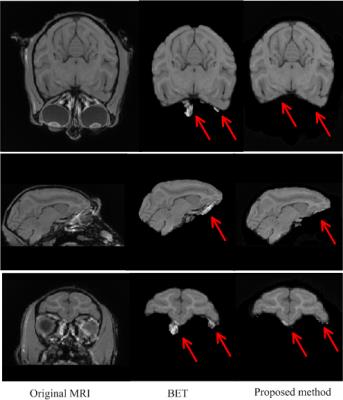
Fig. 2. The comparison of brain extraction results by two methods. The first column is the original MRI. The second and third columns are the results by BET and our proposed method, respectively.
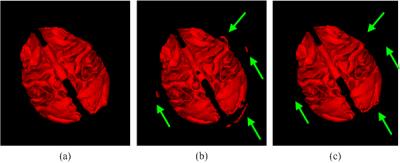
Fig. 3. The comparison of 3D renderings on WM. (a) Ground truth. (b) Conventional method. (c) Proposed method.
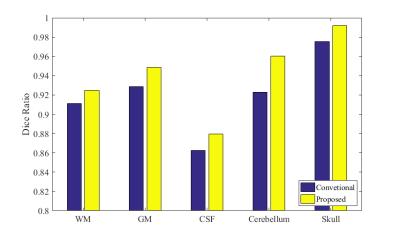
Fig. 4. Comparison of Dice Ratios between our proposed method and the conventional method.