2155
Evaluation of the impact of blood inflow on free-breathing 2D dynamic oxygen-enhanced MRI1Bioxydyn Ltd, Manchester, United Kingdom, 2School of Health Sciences, The University of Manchester, Manchester, United Kingdom
Synopsis
Technical validation of dynamic oxygen–enhanced MRI (OE-MRI) techniques is required for them to become accepted and useful imaging biomarkers. In this work we quantify the impact of using different scanner platforms and protocols on the parameterisation of dynamic single-slice OE-MRI of the lung. Results show that blood in-flow effects consistently provided lower estimates of baseline T1 and higher estimates of maximum change in partial pressure of oxygen, but it does not influence the wash in time estimation. This suggests that sensitivity to variation in ventilation is approximately equivalent using both protocols.
Purpose
Technical feasibility and clinical utility of dynamic oxygen–enhanced MRI (dOE-MRI) techniques has previously been demonstrated [1]. However, the impact of different scanner platforms using different protocols on dOE-MRI results has not yet been quantified. In this work we present results from a multi-vendor free-breathing dOE-MRI comparison study that used 2D single-slice OE-MRI protocols with different sensitivity to blood in-flow effects on two different 1.5 T MRI systems.Methods
8 healthy volunteer (5 females and 3 males, mean age 33.5 years (σ=11.6)) were scanned on two 1.5T systems from different manufacturers on different dates. Off-the-shelf protocols were optimised on both systems to map changes in lung signal due to changes in inhaled oxygen concentration. To maximise T1 weighting and minimise susceptibility artefacts, both protocols used an inversion prepared half-echo fast spin echo (HASTE) readout with centre out phase reordering sequence. System A allowed the use of a non-selective inversion pulse with HASTE readout module, which allows blood flowing into the imaging plane to experience the same magnetisation preparation as the static tissue within the image slice. Due to the software configuration of System B, a non-selective inversion preparation was not available, so instead a slice selective inversion with HASTE readout module was used. In this case, blood flowing into the imaging plane is not inverted and therefore does not experience the same T1 weighting as the static tissue within the image slice.
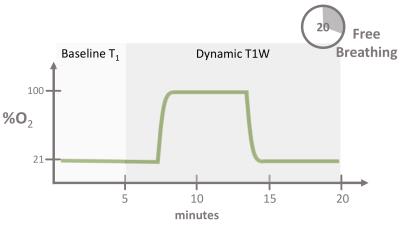
The OE-MRI protocol lasted approximately 20 min on both systems and consisted of baseline variable inversion recovery T1 measurement followed by dynamic acquisition (see figure 1), both acquired during free breathing and without respiratory or cardiac gating. Baseline T1 maps were acquired while subjects breathed medical air (21% O2), followed by 14 min dynamic acquisition (6 s/frame), during which gas was switched from air to 100% O2 and back again to air, as detailed in figure 1. Gas was delivered to the subject at 15 L/min using a non-rebreathing mask (Intersurgical Eco 1181, Berkshire, UK).
Images were registered to correct for breathing motion and changes in the partial pressure of oxygen over time (∆PO2(t)) in the lung parenchyma was derived from the dynamic time series and baseline T1 maps. ∆PO2(t) curves were then parameterised using a mono-exponential function to generate maps of the oxygen wash-in time (τup – related to ventilation) and the maximum change in tissue partial pressure of oxygen (∆PO2max) [2]. All analysis was performed using Pulmolux (www.pulmolux.com).
Results
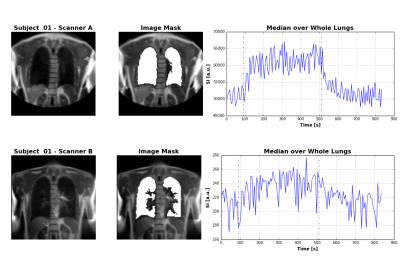
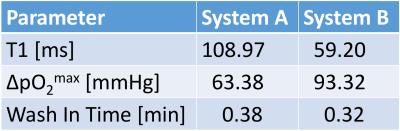
The example dataset in figure 2 shows the similar image contrast achieved on both systems, but also highlights the more pronounced inflow artefact on System B caused by the slice selective inversion, which affects quantification of the oxygen kinetic parameters in the lung. Quantification on both systems was summarised in terms of the median of voxel values over both lungs, shown in figure 3 and their variability was quantified in terms on the interquartile range across the 8 subjects, shown in Table 3. System B consistently provided lower estimates of baseline T1 and higher estimates of ∆PO2max. No significant differences were observed in τup between the groups, except for a single data set that showed very long τup when using system B.Discussion
Lung T1 estimated from System A is closer to literature values at 1.5 T [3]. The lower T1 estimated using system B is due to the slice selective inversion preparation, allowing unperturbed blood to flow into the imaging slice. This effect will be less at short inversion times than at longer inversion times, meaning that the apparent relaxation rate of the signal within the imaging slice appears greater (i.e. lower apparent T1). As the estimated ∆PO2 is proportional to the change in the inverse of T1, the underestimation of baseline T1 appears as an overestimation of ∆PO2max when using System B. However, blood inflow effects do not influence the wash in time estimation, suggesting that this parameter is generally the most robust of the measured kinetic parameters. However, as shown in figures 2 and 3, outlier values of τup can arise due to the noise in the time series caused by variations in inflowing blood.
Conclusions
Differences in off-the-shelf sequences can lead to errors in the parameterisation of dynamic single-slice OE-MRI of the lung. Differences in the sensitivity to blood inflow caused by differences in magnetisation preparation schemes can lead to biased parameter estimates. However, the rate of signal change due to inhalation of elevated levels of oxygen is relatively robust to such errors. This suggests that sensitivity to variation in ventilation is approximately equivalent using both protocols.
Acknowledgements
Thanks to University of Manchester, MRIF, MARIARC and Philips HealthcareReferences
[1] Kruger, S. J., Nagle, S. K., Couch, M. J., Ohno, Y., Albert, M. and Fain, S. B. (2016), Functional imaging of the lungs with gas agents. J. Magn. Reson. Imaging, 43: 295–315. [2] Kershaw, L. E., Naish, J. H., McGrath, D. M., Waterton, J. C. and Parker, G. J. M. (2010), Measurement of arterial plasma oxygenation in dynamic oxygen-enhanced MRI. Magn. Reson. Med., 64: 1838–1842. [3] Kauczor, Hans-Ulrich (Ed.) (2009), MRI of the Lung. Springer.Figures