2131
Gradient tracing for segmentation of low resolution, low T1-weighted breast MR images1Radiology, UW- Madison, Madison, WI, United States, 2Biostatistics and Medical Informatics, UW- Madison, WI, United States, 3Medical Physics, UW- Madison, Madison, WI, United States, 4Medicine, UW- Madison, Madison, WI, United States, 5Biomedical Engineering, UW- Madison, Madison, WI, United States, 6Emergency Medicine, UW- Madison, Madison, WI, United States, 7Carbone Cancer Center, UW- Madison, Madison, WI, United States
Synopsis
Segmentation of breast MR images remains a challenge and a necessity for a variety of quantitative applications. We present a semi-automatic methodology for segmentation of breast tissue for the special case of low resolution, low flip angle chemical shift encoded MRI (CSE-MRI) with water-fat separation. User interaction is required to set the bounds of the segmentation, while the chest wall and skin are segmented automatically. The results differed with corrections by an experienced radiologist by 4.2% average error per case. The method exhibits comparable accuracy to published methods and high agreement between non-expert reviewers.
Purpose
Segmentation of breast MR images is a challenging problem due to high inter-patient anatomic variability (Figure 1), yet is necessary for quantitative assessment of the breast that requires the exclusion of the skin and chest wall. While a number of published methods currently exist1,2,3, most rely on contrast differences between fat, fibroglandular tissue, skin, and muscle tissues to produce reliable segmentation. However, for some quantitative applications4 it may be necessary to limit T1 weighting through the use of a low flip angle which also lowers the SNR and tissue contrast. Additionally, low resolution imaging complicates segmentation due to partial volume effects. The purpose of this work was to demonstrate a novel methodology for segmenting low resolution breast MR images without reliance on T1-weighting for tissue differentiation.Methods
Algorithm
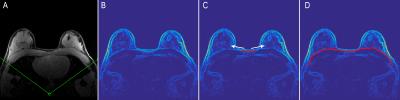
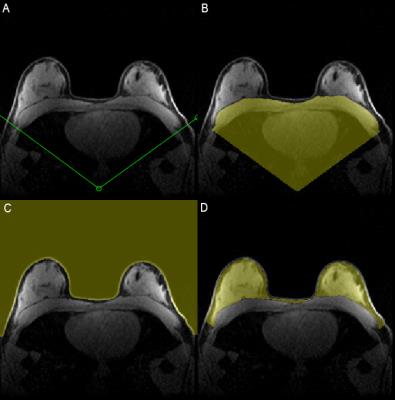
Starting with an axial breast MR image, the lateral boundaries of the breast are designated with a published Vcut method1,2 modified to have the user define the Vcut on the most superior slice where the edges of the pectoralis muscle are distinguishable. The superior and inferior boundaries are defined by the user through visual inspection of the volume in the sagittal plane.
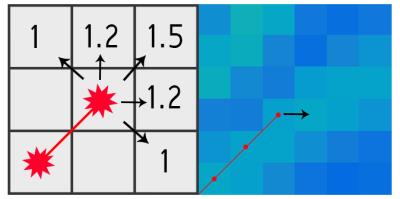
The chest wall is identified automatically by an algorithm that traces the gradient magnitude image. The sternum is detected first on the image as it is the most consistently identifiable landmark of the chest wall. From the sternum, a path is traced laterally one pixel at a time, choosing the largest weighted value of the gradient at the leading edge of the trace (Figure 2). Once the path has reached the lateral boundary of the breast, it terminates. Constraining this trace to a low tortuosity prevents it from deviating far from the true chest wall.
Segmentation starts on the central slice and continues to the selected superior and inferior boundaries. The location of the sternum in the current slice is used to define the starting location for sternum identification in the next slice.
An automatic thresholding operation was used to identify the skin-air boundary. The average skin thickness (pixels) is estimated using the inner edges of the skin using the Canny method5. A border of this thickness is then excluded on the entire skin-air boundary. The overall segmentation strategy is summarized in Figure 4.
Validation
This study was IRB-approved and HIPAA compliant. To validate the method, 50 consecutive patients undergoing routine clinical breast MRI underwent informed consent. Patients with a current or previous diagnosis of breast cancer or breast implants were excluded, resulting in 27 subjects.
Images with a resolution of 1.4x1.4x2.5mm (44 slices, 1° flip angle, 1:15 acquisition time) were acquired using a multi-echo chemical shift encoded (CSE) technique and reconstructed to generate fat/water separated images using a previously described CSE-MRI method4 .
All cases were processed automatically by the algorithm after the user defined boundaries. Once segmented, one fellowship-trained breast radiologist with 7 years of experience and two non-clinical researchers each manually corrected the automated segmentations on a slice-by-slice basis.
Inclusion errors were defined as voxels in which the algorithm selected non-breast tissue, and exclusion errors were defined as voxels in which the algorithm failed to select breast tissue, according to the reviewer. Percent total difference was calculated as the sum of these errors divided by the total segmented volume defined by the radiologist. The corrected segmentations of the radiologist were compared to the uncorrected algorithm as well as to each of the lay reviewers.
Results
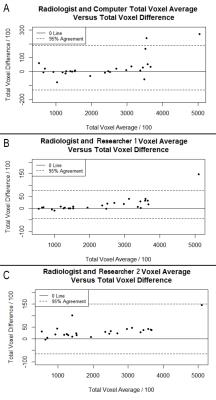
Each case was automatically segmented in less than 10 seconds after selecting user-defined boundaries. The percent total difference between the radiologist and algorithm averaged 4.22% (median: 1.87%, min: 0.23%, max: 19.66%, SD: 5.05%, 95% CI: [2.21%, 6.22%]). Differences were largest at the extremes of breast size and there was excellent agreement between the corrected segmentations performed by the radiologist and two researchers (Figure 5).Discussion
The segmentation algorithm averaged less than 5% total difference per case from an experienced radiologist, similar to that of a previously published method2 despite the challenges of low resolution and low T1-weighting. Additionally, the excellent agreement between the radiologist’s corrected segmentation and that of non-clinical researchers means that for applications requiring refinement of the automated segmentation, reliable results are produced by a non-clinician.Conclusion
We describe a method to segment breast MR images and remove the chest wall and skin with processing time of 10 seconds and mean total difference of 4.2% from an expert radiologist. The method does not require high resolution or significant T1-weighting. Future directions include a self-correcting algorithm, and development of the software for public use.Acknowledgements
The authors would like to acknowledge support from the Department of Radiology at the Authors' institution, the Cancer Center at the Authors' institution, the NIH/NCI (T32 CA009206, P30 CA014520, R01 DK083380, K24 DK102595), the Radiological Society of North America, and GE Healthcare.References
[1] Nie K, Chen JH, Chan S, Chau MK, Yu HJ, Bahri S, Tseng T, Nalcioglu O, Su MY. Development of a quantitative method for analysis of breast density based on three-dimensional breast MRI. Med Phys. 2008 Dec; 35(12):5253-62.
[2] Muqing Lin, Jeon-Hor Chen, Xiaoyong Wang, Siwa Chan, Siping Chen, and Min-Ying Su. Template-based automatic breast segmentation on MRI by excluding the chest region. Medical physics, 40(12):122301, 2013.
[3] Wang, Lei, et al. "Fully automatic breast segmentation in 3D breast MRI." 2012 9th IEEE International Symposium on Biomedical Imaging (ISBI). IEEE, 2012.
[4] Roberta M Strigel, Leah Henze Bancroft, Diego Hernando, and Scott B Reeder. Proton density water fraction as a measurement of breast fibroglandular tissue volume and concentration. Abstract 2480.Singapore, May 2016. International Society for Magnetic Resonance in Medicine 23rd Annual Meeting & Exhibition.
[5] Canny, John, "A Computational Approach to Edge Detection," IEEE Transactions on Pattern Analysis and Machine Intelligence, Vol. PAMI-8, No. 6, 1986, pp. 679-698.
Figures