2105
Reducing computation time for registration in Breast DCE-MRI: Effects of percent sampling on kinetic analysis model parameters, uncertainties, and goodness of Fit1Medical Biophysics, Western University, London, ON, Canada, 2Lawson Imaging, Lawson Health Research Institute, London, ON, Canada, 3Surgical Oncology, London Regional Cancer Program, London, ON, Canada, 4Oncology, Western University, London, ON, Canada, 5Radiation Oncology, London Regional Cancer Program, London, ON, Canada, 6Medical Imaging, Western University, London, ON, Canada, 7Diagnostic Imaging, St. Joseph's Health Center, London, ON, Canada, 8Physics and Engineering, London Regional Cancer Program, London, ON, Canada
Synopsis
Patient movement during dynamic contrast enhanced breast MRI acquisition can degrade signal enhancement curves. Non-rigid image registration can improve enhancement curves, but computation time can be long, especially if all voxels are sampled for cost function estimation. This work investigates the influence of the percentage of voxels sampled (PS) on goodness-of-fit, kinetic model parameter values and uncertainties. The spatial distribution of parameter values was more strongly influenced by registration and PS compared to global tumor measures. Registration with very low PS values increased parameter uncertainties compared to unregistered. 5 PS provided similar performance to 100 PS with reduced computation time.
Introduction
Dynamic contrast enhanced (DCE) MRI is useful to aid diagnosis and treatment monitoring of breast cancer. However, patient movement can introduce artificial variation in the signal enhancement curves. Non-rigid image registration can greatly improve signal enhancement curves1, but computation time can be long. Reducing the percent of voxels sampled (PS) for estimating the cost function could reduce computation time, but may affect accuracy and precision of the registration. This work investigates the influence of PS on kinetic model parameter values, precision, and goodness-of-fit.Methods
DCE-MRI breast images were acquired on a 3T-PET/MRI system (Siemens Biograph mMR) in 8 patients with early stage breast cancer. Three-dimensional fat suppressed fast low angle shot (FLASH) images (spatial resolution=1.0x1.1x2.0mm, time resolution=18s) were acquired from patients prior to and at 28 time points following Gadovist (0.1 mMol/kg) injection. Deformable image registration of the affected breast was performed with 3DSlicer2 on a PC running windows 10 (16GB ram, intel i7-4790 CPU). Mattes mutual information was used as a cost metric with a 2cm isotropic control point spacing. Each post-contrast image was registered to the pre-contrast image, and was repeated with PS values of 0.5%, 1%, 5%, 20%, and 100%. Tumours were segmented using Otsu’s method3. The Toft’s kinetic model4 was fit voxel-by-voxel to segmented images using a population derived AIF5. The root-mean-square-error (RMSE) about the fitted curve, best-fit values, and standard error (σKtrans, σkep) of the model parameters (Ktrans, kep) were extracted from each voxel. Absolute difference maps (ΔKtrans, Δkep) were created by taking the absolute value of voxel-by-voxel differences between 100 PS and other PS cases (including unregistered data) in order to assess changes in the spatial distribution of parameter values with PS.
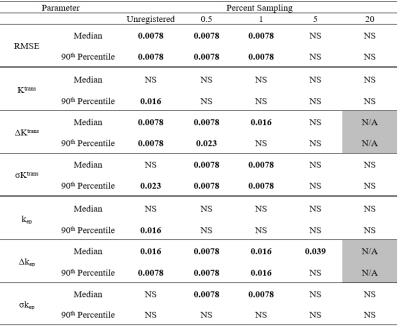
For each parameter (Ktrans, kep, σKtrans, σkep, ΔKtrans, Δkep, RMSE) within each patient image, the medians and 90th percentile over all voxels in the segmented tumour were determined. Pair-wise Wilcoxon rank-sum tests were performed to compare values for 100% PS to those for lower PS and unregistered, except for ΔKtrans and Δkep which were compared to 20 PS since they were subtracted from 100 PS.
Results/Discussion
RMSE values for unregistered and low PS (0.5%, 1%) were significantly greater than 100 PS (Table 1) but not significantly different for 5 and 20 PS, indicating an improvement to the goodness-of-fit with registration at sufficiently high PS (5%).
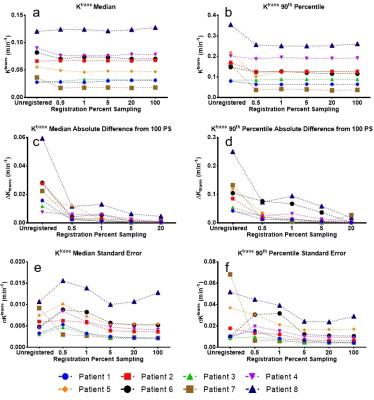
Figure 1a shows that changes in the median Ktrans value appear to be small across all PS cases (including unregistered). Differences were not significant for all comparisons with 100 PS (Table 1). However, 90th percentile Ktrans values (Fig 1b) for unregistered images were significantly greater than those from 100 PS. Findings for kep were similar (Table 1).
Figure 1c and 1d demonstrate that pixel-by-pixel ΔKtrans were more sensitive to image registration than median and 90th percentile values of Ktrans. The differences were significant for unregistered, 0.5 and 1 PS but not 5 PS when compared to 20 PS (Table 1). This trend is reflected in Δkep (figure not shown) except that the median value for 5 PS was significantly different from 20 PS. Changes in ΔKtrans and Δkep indicate changes in the spatial distribution of corresponding model parameters (Ktrans and kep).
Regarding σKtrans (Fig. 1e and f), values for some patients were higher (i.e., higher fitting uncertainty) compared to unregistered or higher PS. This is supported by the statistical differences for σKtrans and the median value of σkep (Table 1). This trend differs from those of ΔKtrans and Δkep which did not increase from unregistered to low PS registered images. We speculate that with registration at low PS, the deviation of the overall DCE curve shapes from that predicted by the Tofts model is reduced (RMSE, Table 1), but residual high frequency fluctuations in the curves leads to high values for σKtrans and σkep.
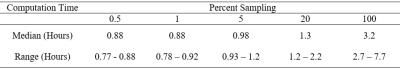
From Table 1, only one significant difference occurred at or above 5 PS. Considering also that any reduction in computation time (Table 2) at PS less than 5 was very small, our results suggest that 5 PS seems to provide a reasonable balance between computation time and registration performance. Limitations of this work include the relatively small number of patients and the possibility that the impact of PS on image registration could depend on imaging parameters such as spatial resolution and signal-to-noise ratio.
Conclusion
For image registration of breast DCE the choice of percent of voxels sampled for estimation of the cost function has an impact on the kinetic analysis derived parameters and computation time. Our results suggest that 5% sampling is a reasonable choice.Acknowledgements
Breast MRI coil courtesy Siemens healthcare Canada.References
[1] Chebrolu VVN, Shanbhag D, Gupta S, Hervo P, Reid S. Impact of Non-Rigid Motion Correction on Pharmaco-Kinetic Analysis for Breast Dynamic Contrast-Enhanced MRI. 23rd ISMRM Annu Meet. 2015.
[2] Fedorov A, Beichel R, Kalpathy-Cramer J, et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging. 2012;30(9):1323-1341. doi:10.1016/j.mri.2012.05.001.
[3] Otsu, N., A Threshold Selection Method from Gray-Level Histograms,"IEEE Transactions on Systems, Man, and Cybernetics, Vol. 9, No. 1, 1979, pp. 62-66.
[4] Tofts PS. Modeling tracer kinetics in dynamic Gd-DTPA MR imaging. Journal of Magnetic Resonance Imaging. 1997 Jan 1;7(1):91-101.
[5] Parker GJM, Roberts C, Macdonald A, et al. Experimentally-derived functional form for a population-averaged high-temporal-resolution arterial input function for dynamic contrast-enhanced MRI. Magn Reson Med. 2006;56(5):993-1000. doi:10.1002/mrm.21066.
Figures