2077
Using ngram-based features to explore the correlation of prostate MR findings and PI-RADS classification1Peking University First Hospital, Beijing, People's Republic of China, 2Philips Research China, Shanghai, People's Republic of China
Synopsis
The decision tree trained on MR descriptions by natural language processing (NLP) method represents a desirable performance in identifying low-risk PI-RADS 2-3 classes with high precision and high-risk PI-RADS 5 class with high recall. From the decision path, several specific features are adopted to make decision and the identification of key indicator contributes to distinguish PI-RADS 2 class from PI-RADS 3 class.
Purpose
Natural language processing (NLP) provides techniques translating natural human language into a structured format, and enables automatic identification and extraction of prostate MR descriptions.1 The objective of this study is to explore the correlation of prostate MR findings and PI-RADS classification by NLP approach.Methods
Data of prostate MR were retrospectively obtained from Hospital Information System (HIS) from Jan. 2014 to Oct. 2016 and stored in Excel spreadsheets. NLP model was constructed and tested in the following steps:
1. Corpus pre-process: The MR diagnosis and PI-RADS scores were extracted from the Excel spreadsheets and stored in a flat txt file for each patient.
2. Feature Generation and Selection: Firstly, normalization step expanded abbreviations to their full form (e.g., “BPH” was replaced by “Benign Prostate Hyperplasia”), which makes it feasible to capture the key information from MR diagnosis by n-grams. The process of word segmentation was followed by the generation of 3-grams and 4-grams. Anatomic sites of prostate (e.g., “peripheral zone”, “central zone” and “seminal vesicle”) were added to the dictionary to improve the accuracy of word segmentation. An n-gram is selected as feature candidate only if it has complete information about the lesion, otherwise those with incomplete semantic meaning were excluded by regular expressions, which were generated based on manual review and summary. Finally, each of the 1899 pieces of MR diagnosis was represented by a vector, including the frequencies of selected features (n-grams). We conducted the decision tree-based classification on this data through 10-fold cross validation (CV) to explore the correlation of MR diagnosis and PI-RADS scores (PI-RADS 1-5).
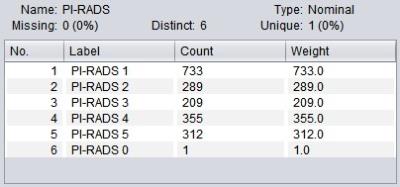
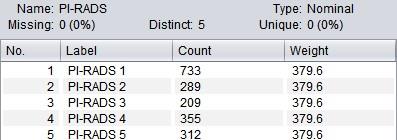
3. Data pre-process: As PI-RADS 0 indicates a relatively low risk and there is only one case in the original data set, this classification was excluded. To deal with the unbalanced class distribution, specific weights were assigned to each class during the model training (Figure 1-2).
Results
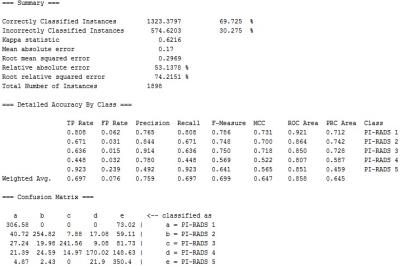
Ngram-based Classification Totally 1898 PI-RADS scores (PI-RADS 1, n=733; PI-RADS 2, n=289; PI-RADS 3, n=209; PI-RADS 4, n=355; PI-RADS 5, n=312) and the corresponding MR findings in the original data set were included. A decision tree was trained to classify the MR diagnosis into one of the 5 PI-RADS classes (i.e. PI-RADS 1-5). The 10-fold cross validation (CV) shown in Figure 4 demonstrates that the decision tree has desirable performance (AUC=0.858), especially a high sensitivity of “PI-RADS 5” class (Recall=0.923) and high precision of relatively low-risk classes (Precision of “PI-RADS 3” and “PI-RADS 2” reaches 0.914 and 0.844, respectively). Looking into the decision tree (Figure 3), we found the size of this tree is modest and the decision can be made based on some specific features, such as “hypointense on T2WI”, “BPH”, “hemorrhage in central zone”, “inflammation”, “possibility of cancer” etc. It should also be noted that “PI-RADS 2” class is distinguished from “PI-RADS 3” by the mention of “further prostate biopsy is recommended”, which is a key indicator to further distinguish PI-RADS classification.Discussion
We have shown that, the decision tree represents a desirable performance in identifying low-risk PI-RADS 2-3 classes with high precision and high-risk PI-RADS 5 class with high recall, furthermore a key indicator is identified to distinguish PI-RADS 2 class from PI-RADS 3 class, which indicates that the MR findings may be similar for these two classes thus the physician would be cautious in this case and leave the final decision to biopsy.Conclusion
It is feasible to use the natural language processing in identifying the high-risk PI-RADS 5 class. Further improvements are necessary to assess the stability and reproducibility of the result and are expected to develop an automatic grading application of PI-RADS.Acknowledgements
No acknowledgement found.References
1. Pons E, Braun LM, Hunink MG, et al. Natural Language Processing in Radiology: A Systematic Review. Radiology. 2016;279(2):329-343.