1862
Exploring Cortical Fiber Crossings in Mice using Diffusional Kurtosis Imaging1Department of Radiology and Radiological Science, Medical University of South Carolina, Charleston, SC, United States, 2Center for Biomedical Imaging, Medical University of South Carolina, Charleston, SC, United States, 3Department of Neurology, Medical University of South Carolina, Charleston, SC, United States, 4Department of Neuroscience, Medical University of South Carolina, Charleston, SC, United States
Synopsis
The ability of diffusional kurtosis imaging (DKI) to detect multiple intravoxel fiber directions in vivo is demonstrated for mouse cortex, with two or more directions being detected in the majority of voxels. The distribution of angular differences between the different fiber directions for individual voxels peaked at near 90o, suggestive of a grid-like pattern of neurites. Our findings support the feasibility of DKI-based tractography in mouse cortex.
Purpose
The non-invasive mapping of neuronal connections in cortical gray matter is challenging with diffusion MRI because the cortex has relatively low diffusional anisotropy, presumably due to an abundance of intravoxel fiber crossings. Here we explored the ability of diffusion MRI to detect multiple fiber directions in mouse cortex by using diffusional kurtosis imaging (DKI). Robust detection of multiple intravoxel fiber directions would demonstrate the potential of DKI-based tractography of neuronal networks in mouse cortex.Methods
Conventional DKI data from a 24 month old male C57bl/6 mouse were collected using a Bruker BioSpec 7T MRI system with 64 diffusion encoding directions and b = 0, 1000, 2000 s/mm2. Coronal images were acquired with an in-plane resolution of 0.23 x 0.23 mm2 and a slice thickness of 0.4 mm. A T2 TurboRARE dataset with the same slice thickness was obtained as an anatomical reference. Diffusion and kurtosis tensors were estimated using the Diffusional Kurtosis Estimator1 (DKE, Medical University of South Carolina, Charleston, USA, http://academicdepartments.musc.edu/cbi/dki/dke.html). Cortical voxels were determined by thresholding FA<0.25 with subsequent semi-automatic removal of deep gray matter. To explore the diffusion patterns, we calculated, in each voxel, a kurtosis diffusion orientation distribution function2 (dODF) using a radial weighting power of four. Gaussian dODFs, which encode the same information as the traditional diffusion ellipsoid, were also calculated from just the diffusion tensor (constructed with only b = 1000 s/mm2), to visualize the difference between resolving for single (Gaussian dODF) or multiple fibers (kurtosis dODF) in the cortex. Fiber directions were estimated from dODF maxima.Results
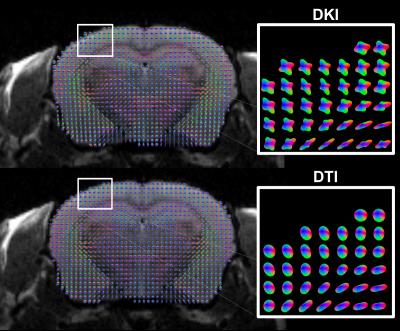
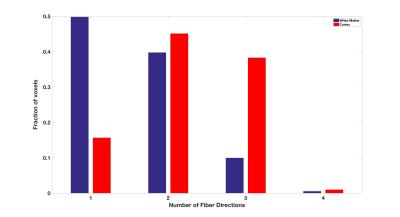
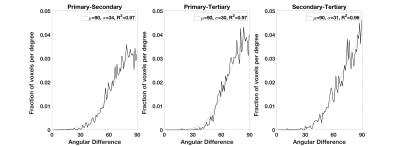
To illustrate fiber crossing detection with DKI, a single coronal slice with superimposed kurtosis dODFs is depicted in Fig. 1. Throughout the mouse cortex, the kurtosis dODF consistently resolves multiple fiber directions. As a reference, also in Fig. 1 are the corresponding Gaussian dODFs, which reflect the ability of conventional diffusion tensor imaging to detect a single, dominant fiber direction but not fiber crossings. A histogram for the number of detected fiber directions in each voxel with the kurtosis dODF from both cortex and white matter is shown in Fig. 2. Whereas a fiber crossing was detected in about 50% of voxels in white matter, in cortical gray matter over 80% contained a fiber crossing. The average number of fiber directions per cortical voxel was 2.3. The distribution of angular differences calculated between the various fiber directions within a given cortical voxel are plotted in Fig. 3. The angular distribution for each pair of fiber directions is reasonably well approximated (average R2 of 0.97) by a half-normal distribution with a peak at 90o and standard deviations of 34o, 30o, and 31o between directions primary-secondary, primary-tertiary, and secondary-tertiary, respectively, where the directions have been ordered according to the magnitude of the corresponding dODF maxima.Discussion
In cortical gray matter, the kurtosis dODFs constructed using DKI detect multiple intravoxel fiber directions in the majority of voxels. On average, we detect more than two fiber directions per cortical voxel. White matter, in contrast, is found to consist half of voxels with only a single fiber direction (Fig. 2), which is similar to previously reported results for human brain2. The distribution of angular differences between the fiber directions within individual voxels is peaked at approximately 90o (Fig. 3), which is in rough accordance to the grid-like geometrical organization proposed by Wedeen, et al3. A similar fiber crossing angle distribution has previously been found with DKI for white matter in human brain2. These observations, together with the qualitative results from Fig. 1, support the feasibility of performing fiber tractography in mouse cortex using DKI. Note that hippocampal gray matter tractography has already been demonstrated in mouse with the alternative approach of constrained spherical deconvolution4.Acknowledgements
This work was supported in part by National Institutes of Health research grants T32DC0014435, T32GM008716, DC014021 and The Litwin foundation.References
1. Tabesh A, Jensen JH, Ardekani BA, Helpern JA. Estimation of tensors and tensor-derived measures in diffusional kurtosis imaging. Magnetic resonance in medicine. 2011;65(3):823-836.
2. Jensen JH, Helpern JA, Tabesh A. Leading non-Gaussian corrections for diffusion orientation distribution function. NMR in Biomedicine. 2014;27(2):202-211.
3. Wedeen VJ, Rosene DL, Wang R, Dai G, Mortazavi F, Hagmann P, Kaas JH, Tseng WY. The geometric structure of the brain fiber pathways. Science. 2012;335(6076):1628-1634.
4. Wu, Dan, Jiangyang Zhang. In vivo mapping of macroscopic neuronal projections in the mouse hippocampus using high-resolution diffusion MRI. NeuroImage.2016; 125: 84-93.
Figures