1844
Harmonization for DTI measurements mapping across sites in multi-center MRI study1Department of Biomedical Engineering, the Key Laboratory of Biomedical Information Engineering of the Ministry of Education, Xi'an Jiaotong University, Xi'an, People's Republic of China, 2Xidian University, Xidian University, People's Republic of China, 3Center for Language and Brain, Shenzhen Institute of Neuroscience, Shenzhen University, shenzhen, People's Republic of China, 4Department of Radiology, the First Affiliated Hospital, Xi’an Jiaotong University, xi'an, People's Republic of China, 5Department of Radiology, the First Affiliated Hospital, Xi'an Jiaotong University, Xi'an, People's Republic of China, 6MR Research China, GE Healthcare, Beijing, People's Republic of China, 7Department of Radiology, Xijing Hospital, Fourth Military Medical University, Xi'an, People's Republic of China, 8Department of Radiology, Xijing Hospital, Fourth Military Medical University, xi'an, People's Republic of China, 9Department of Diagnostic Radiology, Xi’an Children Hospital, Xi'an, People's Republic of China, 10Department of Radiology, Xi’an Gao Xin Hospital, Xi'an, People's Republic of China, 11Department of MRI Diagnosis, Shannxi Provincial People’s Hospital, Xi'an, People's Republic of China
Synopsis
Despite the fact that multi-site diffusion imaging studies are increasingly used to study brain disorders, but it is noteworthy that there are large differences among diffusion measurements from different sites. The current study aimed to confirm the variability and to harmonize data across sites. Our results indicated that not only DTI metrics in human brain within inter-site but also within inter-site changed obviously. Furthermore, a brain voxel-based model was developed to harmonize the DTI metrics and to reduce the deviation compared with reference site data and thus improve the reliability of group analysis in multi-center study.
Introduction
Multi-site diffusion imaging studies are increasingly used to study brain disorders. However, inter-site and inter-scanner variability in the acquired data sets poses a potential problem for joint analysis of diffusion MRI (dMRI) data. It was deemed that there exist large differences between diffusion measurements from different sites. Specifically, the inter-site variability in FA and MD is not uniform over the entire brain, but is tissue specific as well as region specific. Inter-site variability in FA can be up to 5% in major white matter tracts[1]. This study aimed to confirm the variability and to harmonize data across sites.Methods
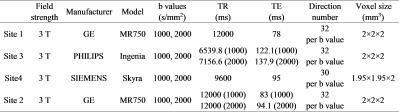
5 healthy young adults (male; age range, 21~23 years) participated in the MR imaging of brain. Each participant underwent one DTI scan in four sites, respectively. This study was approved by the local Institutional Review Board. All parents of participants were informed with the goals and risks of MR scanning and requested written consent before enrollment. In this study, all the scans were performed using 3T MR scanners (site1: MR 750, GE Healthcare; site2: Ingenia, Philips; site3: skyra, Siemens; site4: MR 750, GE Healthcare). Other acquisition parameters were shown in Table 1. The DTI data were automated processed in MATLAB toolbox named “Pipeline for Analyzing brain Diffusion images” ( PANDA). The process mainly included extracting brain images, correcting eddy current, calculating the DTI metrics(FA,MD,AD,RD) and tract-based spatial statistics (TBSS)[2]. We quantified the diffusion MRI variability across sites using reproducibility examined by measures of variability (absolute error relative to the sub and sum about the two scans) across subjects and sites. Then, we harmonized DTI data acquired on scanners of three different manufacturers repeated scanning of 5 participants on GE (site1, site4), Siemens (site2) and PHILIPS (site3) scanners[3]. Specifically, we regarded site1 as the reference group. Then, we inferred a scanner-specific correction factor from the skeletons in different sites and applied this correction factor to the DTI-derived maps in site2, site3, site4. Specifically, skeletons were inferred from the manufacturer-corrected FA maps through TBSS, which enabled a quantitative survey of white matter of the entire brain. In parallel, permutation testing was applied to the DTI metrics derived skeletons.Results
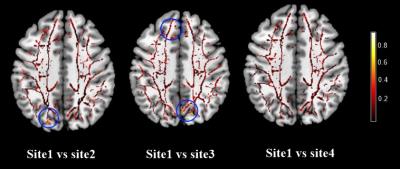
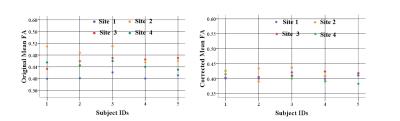
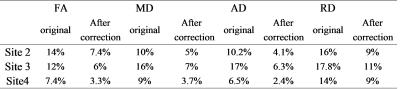
Five male young adults with mean age of 21.8±0.8 of 110.2±9.0 (age range, 21~23 years) were included in this study. The comparisons of FA reproducibility among manufacturers were shown in Fig.1. The results showed significant differences. For the parameters reproducibility between site1 and site4, there were 7% voxels in the skeletons whose FA reproducibility were greater than 0.2, which was less than 12% (site1 vs site2) and 26%(site1 vs site3). The results of other DTI metrics were shown in Table 2. We therefore corrected each map by applying the ratio between two sites with respect to the mean values across the 5 participants: Mean FA site1/ Mean FA site2=0.89; Mean FA site1/ Mean FA site3=0.911; Mean FA site1/ Mean FA site4=0.93. After correction, 3.3% voxels were found that FA reproducibility of that in the skeletons were greater than 0.2 when site1 was compared to site4, while we found that there were 6% in site1 vs site2, and 7.4% in site1 vs site3. Original and corrected means of DTI skeletons for each subjects were shown in Fig.2.Discussion
This study demonstrated the inconsistency of DTI metrics measured. Among four sites, and further suggested the significance of data harmonization across MRI sites. Being consistent with previous study, our results showed that there existed inter-site and inter-scanner variability for joint analysis of dMRI data. Besides, we also can see a smaller variation between sites with the same manufacturer, so it also should be corrected since the nonnegligible variation. Furthermore, to reduce variation within the DTI-based measurements unrelated to factors exclusive of the primary study hypotheses, we applied a harmonization approach to the multi-site data. We obtained good consistency with a good extent (96.7% between site1 and site4; 94% between site1 and site2; 92.6% between site1 and site3 in the skeletons with a FA reproducibility smaller than 0.2. Besides, After correction, mean DTI metrics in the skeletons in site2, site3, site4 substantially drew near to the reference site. Future study is desired to perform a large data-driven robust model to harmonize the DTI data across sites in multicenter study.Conclusion
The variability of DTI metrics in the human brain is obvious across various sites. Harmonizing DTI measurements across scanners would be considered as an effective method to improve the variability across sites and across scanners.Acknowledgements
This study was supported by the National Natural Science Foundation of China (Project Nos. 81371630, 81571752, the Shaanxi Nova program, the Fundamental Research Funds for the Central Universities, National key research and development plan of China, 2016YFC0100300.References
[1] H Mirzaalian, L Ning, P Savadjiev, et al. Inter-site and inter-scanner diffusion MRI data harmonization. NeuroImage. 2016;135:311-323.
[2] Smith, S.M., Jenkinson, M., Johansen-Berg, et al. Tractbased spatial statistics: voxelwise analysis of multi-subject diffusion data. NeuroImage. 2006;31:1487–1505.
[3] Kilian M. Pohl, Edith V.Sullivan, et al. Harmonizing DTI measurements across scanners to examine the development of white matter microstructure in 803 adolescents of the NCANDA study. NeuroImage. 2016;130:194-213
Figures