1842
In Vivo 3D Single-Shot Echo-Planar DWI at 7T for Mapping Tissue Microstructure using Mean Apparent Propagator (MAP) MRI1Center for Neuroscience and Regenerative Medicine, Henry M. Jackson Foundation, Bethesda, MD, United States, 2Radiology and Radiological Sciences, Uniformed Services University of the Health Sciences, Bethesda, MD, United States, 3Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD, United States, 4Center for Neuroscience and Regenerative Medicine, Uniformed Services University of the Health Sciences, Bethesda, MD, United States
Synopsis
A number of advanced diffusion models have demonstrated great promise for mapping tissue microstructure in ex vivo studies with high resolution and fidelity using a wide range of diffusion weightings, but the increased acquisition time (days) is not feasible in vivo. In this study we have addressed some basic DWI acquisition pitfalls by using 3D single-shot EPI on a high-field (7 Tesla) pre-clinical MRI system, and adapted one of the most promising diffusion modeling techniques, mean apparent propagator (MAP) MRI in vivo to derive information about rat brain microstructure within a reasonable time frame.
Introduction
Magnetic resonance (MR) diffusion tensor imaging (DTI) is a non-invasive imaging technique that uses information of magnitude and direction of water molecule diffusion within a voxel to infer features of tissue structure on macro and micro levels.1 Furthermore, DTI appears to be very promising for identifying markers of trauma-related tissue changes following injury.2 In recent years, however, a number of more sophisticated diffusion models have been proposed that may offer improved markers of post-traumatic abnormalities (e.g. DKI, NODDI and MAP-MRI).3-6 It would be advantageous to translate these newer approaches for pre-clinical use in animal models of traumatic brain injury to identify potential biomarkers and test the effects of novel therapeutic strategies.
Diffusion weighted images (DWIs) have poorer image quality and resolution compared to other conventional MR images, especially at very high diffusion weighting, required for newer theoretical methods. DWI artifacts like eddy-current distortion, RF noise, and subject motion, and those arising from the use of faster image acquisition techniques such as echo planar imaging (EPI), including susceptibility-induced distortion, all reduce DWI quality. While more advanced DW techniques show great promise for mapping tissue microstructure in ex vivo studies, 5,6 where high-resolution and high-quality DWIs need to be acquired over a wide range of diffusion weightings, these requirements can increase scan time to several days, which is clearly not feasible for pre-clinical studies and certainly not for clinical translation.
In this study we have addressed some basic DW imaging acquisition pitfalls by using 3D single-shot EPI on a high-field, high-gradient pre-clinical MRI scanner, and adapted one of the most promising diffusion modeling techniques, mean apparent propagator (MAP) MRI 6 in vivo to derive information about rat brain microstructure and cytoarchitecture within a reasonable time frame.
Methods
MRI experiments were conducted using a 7T Bruker
Biospec 70/20 (Bruker Biospin, Billerica, MA) equipped with high-performance
actively shielded gradients (660 mT/m) with integrated shim coils. A birdcage RF
transmit coil in combination with actively decoupled 4-channel RF receive array
coil was used. Four female Sprague‐Dawley rats weighing 250‐300 grams were
scanned 4 times over 10 days to assess repeatability. A single-shot 3D EPI
sequence was used to acquire DWIs: TR=800 ms, TE=40 ms, FOV=22.4×22.4×28.5 mm3,
matrix=128×64×48, resolution=280×280×750 μm3, δ=5 ms and Δ=12 ms. 14 non-collinear
diffusion directions were used for DTI, with b-values=0, 800, and 1600 s/mm2.
For the MAP-MRI acquisition, 32 and 56 diffusion directions were used with b-values=3200
and 4800 s/mm2, respectively. Two sets of data per each diffusion
volume with opposite phase encoding directions (blip-up/blip-down) were
acquired (to correct susceptibility-induced EPI distortion using DR-BUDDI 7
approach). A total of 260 DWI volumes were acquired in vivo within 2 hours. The data were processed using TORTOISE 8
(NICHD, NIH) for DWI corrections and DTI fitting and custom IDL tools for MAP-MRI
fitting according to (6) (implementation of Alan Barnett, NICHD, NIH).
Results and Discussion
There are many practical problems to address when acquiring DWIs for in vivo imaging applications, particularly for MAP-MRI and other methods requiring large b-values. Our goal was to obtain high-SNR high-quality and high b-value DWIs within a reasonable acquisition time. We chose a single-shot 3D EPI (to reduce motion and drift-related artifacts) with 4th-order ghost correction (diminishing the effect related to the oscillation of the readout gradient) to increase SNR and decrease repetition time, providing high-quality DWI data while shortening the overall acquisition time.
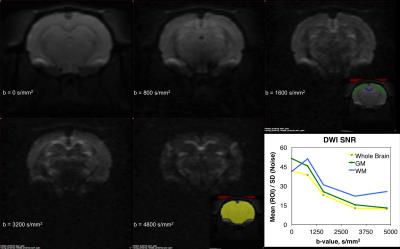
The primary imaging optimizations that were found to be important for obtaining high-quality DWIs were: shimming of imaging region using MAPSHIM (significantly helping with field inhomogeneity), placement of saturation slices to reduce motion artifacts, decreasing the FOV, shortening TE, and measuring the k-space sampling trajectory (Figure 1). Once implemented, we were able to acquire in vivo high-quality and high-SNR DWIs up to b = 4800 s/mm2 with 30 seconds per DWI volume (Figure 2).
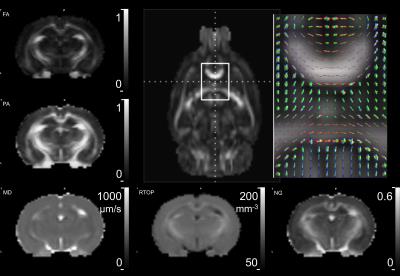
The MAP-MRI modeling framework 6 was used to analyze 260 DWIs from rat brains acquired in vivo using the above-mentioned 3D EPI technique (Figure 3).
Summary
MAP-MRI subsumes DTI while providing new quantitative parameters that reflect intrinsic features of nervous tissue microstructure, in addition to those provided by DTI. For the first time in live animals, MAP-MRI-derived indices were measured and mapped and found to be of high quality, including the propagator anisotropy and non-Gaussianity, which are susceptible to noise and other imaging related artifacts. Results of this study demonstrate the feasibility of migrating a successful ex vivo diffusion MRI acquisition and analysis pipeline to enable in vivo pre-clinical studies.Acknowledgements
This work was funded by the U.S. DOD in the Center for Neuroscience and Regenerative Medicine. PJB and CP were supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development Intramural Research Program.References
1. Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophysical J. 1994; 66(1):259-67.
2. Hulkower MB, Poliak DB, Rosenbaum SB, Zimmerman ME, Lipton ML. A decade of DTI in traumatic brain injury: 10 years and 100 articles later. AJNR Am J Neuroradiol. 2013; 34(11):2064-74.
3. Rosenkrantz AB, Padhani AR, Chenevert TL, Koh DM, De Keyzer F, Taouli B, Le Bihan D. Body diffusion kurtosis imaging: Basic principles, applications, and considerations for clinical practice. J Magn Reson Imaging. 2015; 42(5):1190-202.
4. Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 2012; 61(4):1000-16.
5. Descoteaux M, Deriche R, Le Bihan D, Mangin JF, Poupon C. Multiple q-shell diffusion propagator imaging. Medical Image Analysis 2011; 5(4):603-21.
6. Özarslan E, Koay CG, Shepherd TM, Komlosh ME, Irfanoglu MO, Pierpaoli C, Basser PJ. Mean apparent propagator (MAP) MRI: a novel diffusion imaging method for mapping tissue microstructure. Neuroimage 2013; 78:16-32.
7. Irfanoglu MO, Modi P, Nayak A, Hutchinson EB, Sarlls J, Pierpaoli C2. DR-BUDDI (Diffeomorphic Registration for Blip-Up blip-Down Diffusion Imaging) method for correcting echo planar imaging. distortions. Neuroimage 2015; 106:284-99.
8. Pierpaoli C, Walker L, Irfanoglu MO, Barnett A, Basser P, Chang L-C, Koay C, Pajevic S, Rohde G, Sarlls J, and Wu M. TORTOISE: an integrated software package for processing of diffusion MRI data. ISMRM 18th annual meeting, 2010, Stockholm, Sweden, #1597
Figures