1785
Predicting patient survival in hepatocellular carcinoma (HCC) from diffusion weighted magnetic resonance imaging (DW-MRI) data using neural networks1Image-Based Biomedical Modeling Group, Technical University of Munich, Munich, Germany, 2Institute of Radiology, Technical University of Munich, Munich, Germany, 3Department of Neurology, Ludwig Maximilian University of Munich
Synopsis
In this work we present a method to predict patient survival in hepatocellular carcinoma (HCC). We automatically segment HCC from DW-MRI images using fully convolutional neural networks. In a second step we predict patient survival rates by calculating different features from ADC maps. We calculate Histogram features, Haralick features and propose new features trained by a 3D Convolutional Neural Network (SurvivalNet). Applied to 31 HCC cases, SurvivalNet accomplishes a classification accuracy of 65% at a precision and sensitivity of 64% and 65% when trained using our automatic tumor segmentation in a fully automatic fashion.
INTRODUCTION
Automatic non-invasive assessment of hepatocellular carcinoma (HCC) has the potential to identify subgroups in this highly heterogeneous tumor entity. Subgroup identification could provide a rationale for selected therapies, thus improving patient outcome. In this work we present a novel framework to automatically classify HCC lesions from DW-MRI data.METHODS
We predict HCC malignancy in two steps: As a first step we automatically segment HCC lesions using cascaded fully convolutional neural networks (CFCN) in DW-MRI. A 3D neural network (SurvivalNet) then predicts the HCC lesions' malignancy from the HCC segmentation. We formulate this task as a classification problem with classes being "low risk" and "high risk" represented by longer or shorter survival times than the median survival.
31 Patients underwent clinical assessment and MR imaging for the primary diagnosis of HCC. Barcelona Clinic Liver Cancer Classification was used to assess the clinical stage of the disease. Patients with a history of prior malignancy, insufficient quality due to breathing artifacts, excessive banding or distortion, diffuse tumor growth or non-detectability of the lesions in the DW-MRI sequences were excluded. Imaging was performed using a 1.5 T clinical MRI scanner (Avanto, Siemens) with a standard imaging protocol. Diffusion weighted imaging was performed using a slice thickness of 5mm and a matrix size of 192 by 192.
To automatically detect and segment tumor lesions we applied a cascaded fully convolutional neural network for segmentation as proposed by [1]. We used the DW-MRI as input to the neural network with manual tumor segmentations as training data, which were identified by an experienced radiologist in the early arterial phase and in DWI images (b=600).
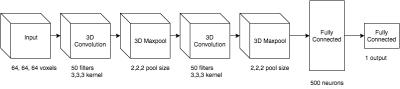
To predict the survival rate of HCC tumor patients we calculated different features using the detected and segmented tumor lesions applied in the ADC image sequences. We calculated handcrafted features such as Histogram features including mean, median, kurtosis and skewness as well as 3D Haralick features [2]. For handcrafted features, we trained a k-nearest-neighbour classifier with k=4 and validated the results using 10-fold cross-validation. Furthermore, we propose new features trained end-to-end by a 3D Convolutional Neural Network (SurvivalNet). The SurvivalNet network architecture is depicted in figure 2.
RESULTS
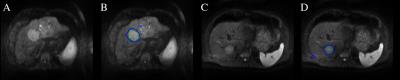
The results of the automatic segmentation are depicted in figure 1. The complex and heterogeneous shape of the tumor lesions was detected and segmented in both images using our automatic segmentation algorithm. The trained model achieves a Dice overlap score of 69% with a specificity of 91%, i.e. almost all lesion voxels are classified as such but false-positive outliers within the liver reduce the overall accuracy and Dice score.
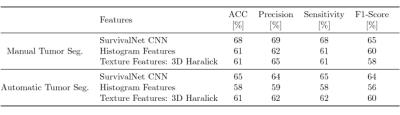
Table 1 shows the quantitative results of our proposed automatic survival prediction framework. SurvivalNet achieves higher scores with both manual as well as automatic segmentation as input compared to handcrafted features. SurvivalNet trained on manual segmentations achieves an accuracy of 68% with a precision and sensitivity of 69% and 68% respectively. Furthermore, SurvivalNet accomplishes a classification accuracy of 65% at a precision and sensitivity of 64% and 65% when trained using our automatic tumor segmentation in a fully automatic fashion. We calculated a paired Wilcoxon signed-rank test with H0: the output posterior class probabilities of SurvivalNet with manual and automatic segmentation belong to the same distribution. At p>0.953, we found H0 to be confirmed, i.e. SurvivalNet produces the same results with automatic segmentation or manual segmentation.
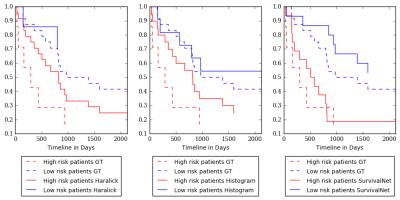
Figure 4 shows a Kaplan-Meier Survival Analysis for Histogram, Haralick and SurvivalNet features. The SurvivalNet is able to split the HCC patients into high risk i.e. short survival and low risk i.e. long survival in contrast to Histogram and Haralick features.
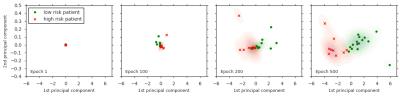
We performed a HCC cluster analysis of the SurvivalNet feature training in figure 3. With increasing training time (from left to right) the SurvivalNet features split the high (red) and low (green) risk HCC cases into distinct clusters. The 500 dimensional SurvivalNet feature space was visualized in this analysis using a Principal Component Analysis (PCA) by showing the two main principal components.
DISCUSSION AND CONCLUSION
The predictive value of various imaging parameters has previously been suggested in HCC [3,4]. In comparison to prior work, we have presented a fully automatic framework to predict survival times of HCC patients. This approach based on fully and 3D convolutional neural networks outperformed state-of-the art handcrafted features, while still achieving the same diagnostic outcome as if human expert segmentations were provided. This work may have potential applications in HCC treatment planning.Acknowledgements
No acknowledgement found.References
[1] Patrick Ferdinand Christ, Mohamed Ezzeldin A. Elshaer, Florian Ettlinger, Sunil Tatavarty, Marc Bickel, Patrick Bilic, Markus Rempfler, Marco Armbruster, Felix Hofmann, Melvin D’Anastasi, Wieland H. Sommer, Seyed-Ahmad Ahmadi, and Bjoern H. Menze, “Automatic liver and lesion segmentation in ct using cascaded fully convolutional neural networks and 3d conditional random fields,” in MICCAI, pp. 415–423. 2016.
[2] R. M. Haralick, “Statistical and structural approaches to texture,”Proceedings of the IEEE, vol. 67, no. 5, pp. 786–804, May 1979.
[3] Wu Zhou, Lijuan Zhang, Kaixin Wang, Shuting Chen, Guangyi Wang, Zaiyi Liu, and Changhong Liang, “Malignancy characterization of hepatocellular carcinomas based on texture analysis of contrast-enhanced mr images,” Journal of Magnetic Resonance Imaging, 2016.
[4] Irina Heid, Katja Steiger, Marija Trajkovic-Arsic, Marcus Settles, Manuela R Eßwein, Mert Erkan, Jorg Kleeff, Carsten Jager, Helmut Friess, Bernhard Haller, Andreas Steingotter, Roland M Schmid, Markus Schwaiger, Ernst J Rummeny, Irene Esposito, Jens T Siveke, and Rickmer Braren, “Co-clinical assessment of tumor cellularity in pancreatic cancer,” Clinical Cancer Research, 2016
Figures