1767
Fast Linear Fitting of Bi-Exponential Intra-Voxel Incoherent Motion (IVIM) Models1Biosciences, SRI International, Menlo Park, CA, United States, 2Psychiatry and Behavioral Sciences, Stanford University, Stanford, CA, United States, 3Bioscience, SRI International, Menlo Park, CA, United States
Synopsis
This work introduces an extremely fast (whole image in ~5s) IVIM fitting procedure based on two sequential linear fits and demonstrates that it is comparable to the more traditional but much slower non-linear fitting. This method is valuable because current fitting methods typically take a significant amount of time, typically from minutes to hours, due to their iterative and non-linear nature. Therefore, this technique can be used as a fitting method alone and also as a way to seed more advanced techniques with accurate starting values.
Purpose
Intra-voxel incoherent motion (IVIM) can probe both diffusion and flow by using a range of b-values, typically 0 to <1000 (s/mm2)1–3. Lower b-values are primarily sensitive to capillary-level flow, and higher b-values are sensitive to tissue diffusion. This ability to measure both diffusion and flow in a single scan is desirable for studies where both metrics are needed, such as in evaluating the effects on the brain of alcoholism4,5 or stroke3. Normally, IVIM data are fitted in multiple stages1,2 and common approaches use nonlinear fitting1,3,6,7. However, these approaches are often too slow to be of clinical use, taking from minutes to hours. This work introduces a fast IVIM fitting procedure (whole 3D image in ~5s) based on two sequential linear fits and demonstrates that it is comparable to the more traditional non-linear fitting.Theory
Equation 1 shows the IVIM signal equation assuming flow (fe-bD*) and diffusion ((1-f)e-bD) compartments. The fitted parameters are S0 (signal), f (perfusion fraction), D* (pseudo-diffusion coefficient), and D (diffusion coefficient) where S(b) is the signal intensity at each of the diffusion values b.
Equation 1: $$S\left(b\right)=S0\left(fe^{-bD^{*}}+\left(1-f\right) e^{-bD} \right)$$
Methods
Subjects:
Healthy normal volunteers (9 men, 6 women; age 23.5 - 67.6 years, mean±sd=41.7±17.4) were scanned on a GE MR750 3T (GE Medical Systems, Waukesha, WI, USA) with an 8-channel head coil (Invivo Co, Gainesville, FL, USA) and with full IRB approval.
Acquisition:
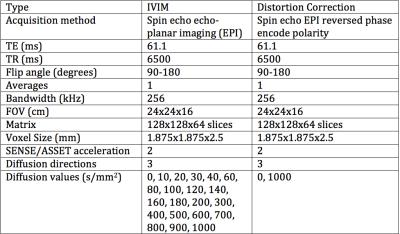
The protocol contained an IR-SPGR, two identical echo-planar imaging (EPI) IVIM sequences, and a reversed phase encode polarity EPI scan (Table 1). Seven subjects were scanned twice with the same protocol >24 hours apart resulting in 44 separate IVIM acquisitions.
Pre-Processing:
All IVIM scans were EPI-distortion and eddy-current corrected using FSL5 (Topup, Eddy)6,8. The SRI24 atlas9 with 116 regions was registered to the IVIM image via the IR-SPGR image using ANTS10. SRI24 brain and parcellation masks were transformed to IVIM space and eroded using a 3x3x3 kernel to minimize cross-region and fluid-tissue contamination.
Non-Linear IVIM Fitting:
This method is described elsewhere11, but the fundamentals are that of a two-step fitting where the parameters are guessed using the high and low b-value data separately1,2 followed by a re-fit using the entirety of the data with the complete bi-exponential model.
Linear IVIM Fitting:
Each region was fit for S0, f, D, D* with S0’, S0* as intermediate variables.
1. Linearly fit ln(S(b>200)) for S0’ and D, where S0’ is the zero intercept of the diffusion fit at b>200.
2. Calculate the residuals for the low b-value data (b<=200) from the fit (data - predicted from step 1).
3. Linearly fit ln(residuals) for S0* and D*, where S0* is the zero intercept of the diffusion fit of the residuals.
4. Calculate S0: S0=S0’+S0*
5. Calculate f as the ratio of the flow signal to the total signal: f=S0*/S0
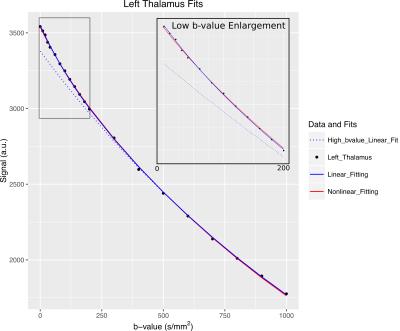
Both fitting procedures were implemented in Python2.712 and produced a single set of IVIM parameters individually for each ROI (Figure 1).
Analysis:
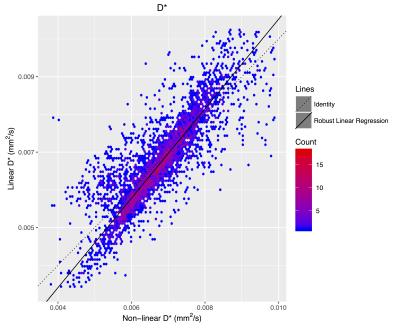
The data cleanup in R13 removed nonlinear fits where the region was smaller than 500 mm3 and non-physical values. No cleanup was performed on the matching linear fits. This resulted in 4233 regions fitted with both methods. The best fit regression line on the non-linear versus linear data was calculated using robust regression in order to most accurately capture the overall trend between the fits.
Results
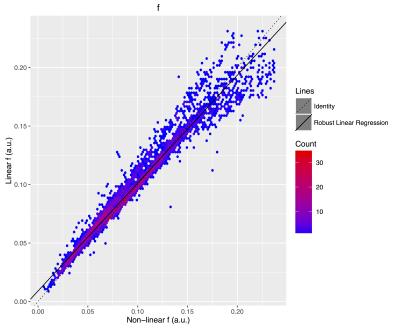
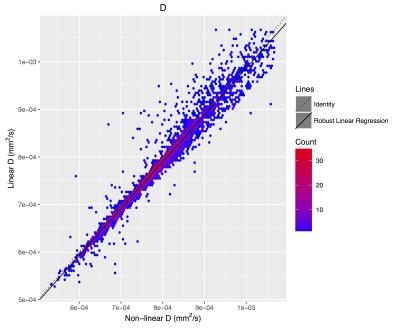
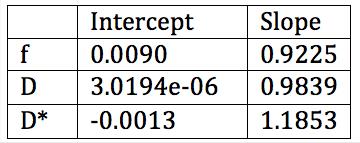
The f (Figure 2), D (Figure 3), and D* (Figure 4) linear fitting values compare very favorably to the nonlinear values (Table 2).Discussion
Linear fitting takes ~5s to complete for the full 3D IVIM image. This fitting method, while simple, is fast, robust, and compatible with more advanced linear fitting methods such as robust or tensor fitting. Additionally, this method can be used as a fast and accurate starting point for more advanced fitting, such as nonlinear or Bayesian algorithms.
Note that this method is similar to the original method proposed by Le Bihan et al.2. However, a major difference is that rather than approximating the combined flow and diffusion compartments with a single exponential, the effect of the diffusion compartment is removed from the data before the flow-only compartment is fit.
The weakness of the proposed method is that it is dependent on the b-value cutoff for the initial fit. This is unavoidable in algorithms that use separate high b-value fitting.
Conclusion
The IVIM metrics are very similar between the traditional nonlinear fit and the proposed linear method. The similarity holds for estimates of f, D, and D*, across a wide range of parameter values and across a large number of brain regions.Acknowledgements
NIH grants: AA021697, AA021695, AA021692, AA021696, AA021681, AA021690, AA021691, K05 AA017168.References
1. Federau C, Maeder P, O’Brien K, Browaeys P, Meuli R, Hagmann P. Quantitative Measurement of Brain Perfusion with Intravoxel Incoherent Motion MR Imaging. Radiology. 2012;265(3). doi:10.1148/radiol.12120584.
2. Le Bihan D, Breton E, Lallemand D, Aubin M-L, Vignaud J, Laval-Jeantet M. Separation of diffusion and perfusion in intravoxel incoherent motion MR imaging. Radiology. 1988;168(2):497-505. doi:10.1148/radiology.168.2.3393671.
3. Federau C, Sumer S, Becce F, et al. Intravoxel incoherent motion perfusion imaging in acute stroke: initial clinical experience. Neuroradiology. 2014:629-635. doi:10.1007/s00234-014-1370-y.
4. Gazdzinski S, Durazzo T, Jahng G-H, Ezekiel F, Banys P, Meyerhoff D. Effects of chronic alcohol dependence and chronic cigarette smoking on cerebral perfusion: a preliminary magnetic resonance study. Alcohol Clin Exp Res. 2006;30(6):947-958. doi:10.1111/j.1530-0277.2006.00108.x.
5. Bühler M, Mann K. Alcohol and the human brain: A systematic review of different neuroimaging methods. Alcohol Clin Exp Res. 2011;35(10):1771-1793. doi:10.1111/j.1530-0277.2011.01540.x.
6. Andersson JLR, Skare S. A model-based method for retrospective correction of geometric distortions in diffusion-weighted EPI. Neuroimage. 2002;16(1):177-199. doi:10.1006/nimg.2001.1039.
7. Orton MR, Collins DJ, Koh D-M, Leach MO. Improved intravoxel incoherent motion analysis of diffusion weighted imaging by data driven Bayesian modeling. Magn Reson Med. 2014;71(1):411-420. doi:10.1002/mrm.24649.
8. Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23 Suppl 1:S208-19. doi:10.1016/j.neuroimage.2004.07.051.
9. Rohlfing T, Zahr NM, Sullivan E V., Pfefferbaum A. The SRI24 multichannel atlas of normal adult human brain structure. Hum Brain Mapp. 2009;31(5):798-819. doi:10.1002/hbm.20906.
10. Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54(3):2033-2044. doi:10.1016/j.neuroimage.2010.09.025.
11. Peterson ET, Zahr NM, Kwon D, Serventi M, Hardcastle C, Sullivan E V. Intra Voxel Incoherent Motion (IVIM) in Brain Regions: a Repeatability and Aging Study. In: Intl. Soc. Mag. Reson. Med. 24. Vol ; 2016.
12. Van Rossum G. Python 2.7, Python Software Foundation. 1991.
13. R Core Team. R Language Definition V. 3.1.1. 2014;3.1.1:55. http://mirror.fcaglp.unlp.edu.ar/CRAN/doc/manuals/r-patched/R-lang.pdf\nhttp://cran.r-project.org/doc/manuals/r-release/R-lang.pdf.
Figures