1753
In-vivo whole-brain Neurite Orientation Dispersion and Density Imaging at sub-millimeter scale using gSlider-SMSElda Fischi-Gomez1,2, Susie Y. Huang1,2, Hui Zhang3, and Kawin Setsompop1,2
1Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital. Department of Radiology, Charlestown, MA, United States, 2Harvard Medical School, Boston, MA, United States, 3Department of Computer Science & Centre for Medical Image Computing, University College London, United Kingdom
Synopsis
We demonstrate the feasibility of applying microstructural models of diffusion to sub-millimeter isotropic resolution whole brain images in-vivo. The NODDI model was successfully fitted to a 700µm DWI dataset acquired using the generalized SLIce Dithered Enhanced Resolution Simultaneous MultiSlice (gSlider-SMS) acquisition1. We demonstrate the ability to map finer-scale structures using this high-resolution data when compared to traditional multi-shell 2 mm isotropic acquisition.
Purpose
Microstructure imaging aims to map important histological properties of tissue non-invasively and in vivo. The current generation techniques are typically based on diffusion-weighted MRI (DWI), which infers tissue microstructure based on its influence on water mobility. These techniques allow in-vivo acquisition at a large number of diffusion directions and multiple b-values, providing high enough signal-to-noise ratio (SNR) to enable the microstructural fitting. Yet, the information inferred from these acquisitions, restricted to the spatial resolution of the image, is inadequate for the study of fine-scale structures. Improved image resolution has the potential to resolve small brain structures and enable the investigation of fine neuroanatomical features. A major hurdle to performing sub-millimeter isotropic resolution dMRI is the inherently low SNR due to the extremely small isotropic voxel sizes especially for DWIs acquired with high b-values. In this work, we demonstrate the feasibility of fitting the NODDI microstructural model at sub-millimeter resolution using the recently proposed gSlider-SMS acquisition approach on the 3T CONNECTOM system.Theory
The gSlider-SMS technique2 enables the acquisition of high-resolution whole-brain DWI. This technique combines Blipped-CAIPI3 and thick-slice super-resolution (SR) techniques4 to enable an SNR efficient high-resolution diffusion acquisition that can acquire data from 10 imaging slices simultaneously. To avoid the blurring due to the inherent ill conditioning of the thick-slice SR reconstruction problem, the excitation RF pulse is modified so that each thick slice has a specially-designed non-uniform phase profile along the slice dimension, which serves as an additional form of RF encoding. This RF phase encoding substantially reduces the coherence between the shifted overlapping thick-slice encoding functions, improving the conditioning of the inverse problem and enables higher-fidelity reconstruction. While gSlider-SMS is SNR-efficient, the extremely small isotropic voxel size targeted means that SNR is still a limiting factor for DWIs acquired with multiple high b-values, needed for multi-compartment models fit. In this work, we choose to combine gSlider-SMS acquisition with the NODDI model, where the minimum affordable SNR for reliable fitting is around 20dB for the non-diffusion weighted DWI acquisition and at least 5dB at high b-values. Voxel size, b-values and number of diffusion directions of the gSlider acquisition were selected to account for these considerations.Methods
One volunteer underwent whole-brain gSlider-SMS acquisition at 700µm isotropic resolution over a 184×314×230 mm3 FOV. The protocol included 64 DWIs and 7 intersperse b=0 images at b=1000 and b=2000 s/mm2, and 122 DWI with 13 intersperse b=0 images at b=3,000 s/mm2. Thick slices (5× larger than nominal slice resolution) with 5 different RF encoding pulses, a multiband factor of 2, Rinplane= RzoomxRgrappa=1.67x2 =3.3x and 6/8ths partial Fourier (PF) encoding were used. The total acquisition time was ~2 hours. The thick slices were reconstructed using slice-GRAPPA reconstruction along with POCS to fill in the missing PF data. A forward model based RF-encoding reconstruction to thin-slice high resolution data was performed [1]. For comparison, the same subject was also scanned in a separate session using at 2-mm isotropic resolution with 64 DWIs and 4 interspersed b=0 images at b=1650, 2550 and 3000 s/mm2. T1w-MPRAGE images were acquired. They were registered to the diffusion space and used to extract white matter and gray matter masks5. DWIs were corrected for eddy currents6 and fitted to the NODDI model using the AMICO framework7 to extract the intra-cellular volume fraction (ICVF), the Orientation Dispersion Index (ODI) and the isotropic volume fraction (isoVF).Results
Figures 1 and 2 show the ICVF and ODI maps obtained after NODDI fitting for the WM and GM, respectively. Figure 3 shows the comparison of the ICVF fitting at 700 µm and 2 mm isotropic resolution. The increased resolution enables investigation of fine neuroanatomical features.Discussion
We demonstrate for the first time the feasibility to image both neurite density and orientation dispersion over the whole brain at sub-millimeter resolution in-vivo. The gSlider-SMS acquisition strategy maximizes SNR over conventional acquisitions due to the thick slab acquisition and provides sufficient SNR for reasonable NODDI fits within the WM. The increased resolution and microstructural information together promise to provide improved in-depth analysis of fine WM structures. The standard NODDI models appear sub-optimal for fitting GM signal at high resolution. Yet our analysis shows the potential utility of gSlider-SMS and NODDI to study the cortical surface. Along with careful analysis of partial volume effect, future work will focus on optimizing this model to improve the quality of obtainable GM microstructure fitting, in order to investigate microstructural features of the cortical/subcortical interface.Acknowledgements
This work was supported in part by the following NIH grants: P41EB015896, U01MH093765, R01EB020613 and R24MH106096 to KS; NINDS K23NS096056 to SYH and by the Swiss National Science Fundation for Research (SNSF) grant P2ELP2_172286 to EFG.References
[1] K. Setsompop, J. Stockmann, Q. Fan, T. Witzel, L. L. Wald. “Generalized SLIce Dithered Enhanced Resolution Simultaneous MultiSlice (gSlider-SMS) to increase volume encoding, SNR and partition profile fidelity in high-resolution diffusion imaging.” Proc ISMRM 2016. [2] K. Setsompop, B. Bilgic, A. Nummenmaa, Q. Fan, S. F. Cauley, S. Huang, I. Chatnuntawech, Y. Rathi, T. Witzel, L. L. Wald. “SLIce Dithered Enhanced Resolution Simultaneous MultiSlice (SLIDER-SMS) for high resolution (700 um) diffusion imaging of the human brain.” Proc ISMRM 2015, p. 339. [3] K. Setsompop, B. A. Gagoski, J. R. Polimeni, T. Witzel, V. J. Wedeen, L. L. Wald. “Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty.” Magn Reson Med 67:1210-1224, 2012. [4] H. Greenspan. “Super-resolution in medical imaging.” Comput J 52:43-63, 2009. [5] Freesurfer toolbox: https://surfer.nmr.mgh.harvard.edu/fswiki [6] FSL toolbox: http://fsl.fmrib.ox.ac.uk/fsl [7] A. Daducci, E.J. Canales-Rodrigues, H. Zhang, T. Dirby, D.C. Alexander and J.-P. Thiran. “Accelerated microstructure imaging via convex optimization (AMICO) from diffusion MRI data.” NeuroImage 105:32-44, 2015.Figures
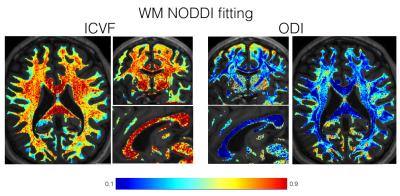
Figure 1: ICVF and ODI maps obtained after the NODDI model
fitting within the WM at 700 μm resolution.
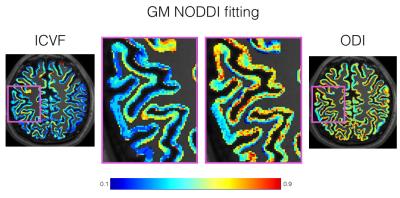
Figure 2: ICVF and ODI maps obtained after the NODDI model
fitting within the GM at 700 μm resolution.
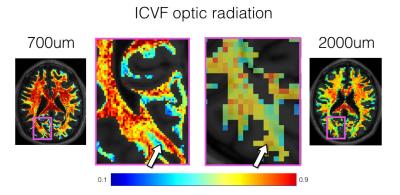
Figure 3: Comparison of the ICVF fitting for the optic
radiation at 700 µm (left) and 2mm (right) isotropic resolution. The increased resolution
enables investigation of fine neuroanatomical features such as
the optic radiations.