1752
Population averaged ferret brain templates for in-vivo and ex-vivo anatomical and diffusion tensor MRIElizabeth B Hutchinson1,2,3, Susan Schwerin3,4, Kryslaine Radomski4, Neda Sadeghi1,2, Michal Komlosh2,3, Jeffrey Jenkins1,2,3, Okan Irfanoglu1,2,3, Sharon Juliano4, and Carlo Pierpaoli1,2
1Quantitative Medical Imaging Section, NIBIB, NIH, Bethesda, MD, United States, 2SQITS/NICHD, NIH, Bethesda, MD, United States, 3Henry M. Jackson Foundation, Bethesda, MD, United States, 4APG, Uniformed Services University of the Health Sciences, Bethesda, MD, United States
Synopsis
In-vivo and Ex-vivo anatomical MRI and DTI templates were generated for the ferret brain along with region of interest segmentation masks based on known ferret neuroanatomy. Templates were built from multiple ferret brain images for each modality using advanced template building tools including symmetric normalization transformation for structural templates and Diffeomorphic Registration for Tensor Accurate AlignMent of Anatomical Structures (DRTAMAS) for DTI templates. The resulting templates are made available on an interactive web site (mriferretatlas.nichd.nih.gov).
Purpose
The main goal of this study was to build and make available MRI and DTI templates for the ferret brain as this is a well-suited species for pre-clinical MRI studies with folded cortical surface, relatively high white matter volume and body dimensions that allow in-vivo imaging with pre-clinical MRI scanners. Four ferret brain templates were built in this study – in-vivo MRI and DTI and ex-vivo MRI and DTI – using brain images across many ferrets as well as region of interest (ROI) masks corresponding to established ferret neuroanatomy[1]. The templates and masks were also used to build a web-based viewer to provide an accessible annotated resource for ferret brain anatomy and to facilitate an open forum for contributions and modifications from others in the community.Methods
In-vivo MRI scans were collected in healthy adult male ferrets including T2W MRI (n=26) and DTI (n=12) using a Bruker 7T horizontal bore small animal MRI system. Multi-echo T2 data were acquired with TE/TR=12-140ms/8-10s and isotropic resolution of 500 microns. DWIs were collected with TE/TR=40ms/5s, 3 b=0 images 32 non-colinear DWIs each for b=700 and 1000 s/mm2. DWIs were collected with 4 total repetitions, 2 “blip-up” and 2 “blip-down”. For ex-vivo image acquisition, 8 perfusion fixed and rehydrated ferret brains were imaged using a 7T Bruker vertical bore microimaging system and 25mm RF coil. Multiple echo T2-weighted MRI was collected using a 3D MSME pulse sequence with TE/TR=10-100ms/3s and isotropic voxel dimensions of 250 microns. For DTI, 88 DWIs were acquired with b=100-3800 s/mm2 and the same spatial geometry and dimensions as the T2 MRI using a 3D EPI pulse sequence with TE/TR=36/700 ms, 8 segments and “blip-up-blip-down” repetitions. DTI processing for in-vivo and ex-vivo DWIs was performed using TORTOISE software[2] to correct for motion and eddy current artifacts as well as geometric distortions[3] and to fit the diffusion tensor. Multiple echo T2 templates for both in-vivo and ex-vivo MRIs were generated using ANTs software tools [4] for template building according to the overview in figure 1 and the diffusion tensor templates for in-vivo and ex-vivo data were generated using Diffeomorphic Registration for Tensor Accurate AlignMent of Anatomical Structures DRTAMAS [5] tools according to the overview in figure 2. A single volume containing 61 labeled masks was generated using the ex-vivo DTI templates and a combination of automated and manual segmentation tools in ITKsnap to define anatomically relevant divisions of the ferret brain according to established ferret neuroanatomy [1]. Additionally, an interactive, publicly available website (mriferretatlas.nichd.nih.gov) has been developed to host and enable users to explore templates and ROI masks described in this work. This web-based viewer contains both a tri-planar MRI and a 3D volume viewer that can be used in conjunction with an annotation capability for several In-vivo and Ex-vivo DTI maps and T2 acquisitions.Results and Discussion
Ex-vivo and in-vivo MRI ferret templates were generated with a range of T2 weightings (figure 3). Major anatomical regions can be delineated by both templates, although the ex-vivo template reveals considerably finer detail. Both templates demonstrate high contrast edges between tissue types which is advantageous for use as a registration target. In-vivo and ex-vivo DTI templates (figure 4) were generated using the recently developed DRTAMAS approach to combine scalar and tensor information to improve both global registration and local fiber alignment for white matter tracts[5]. This resulted in DTI templates with excellent localization of both gray and white matter anatomical features across all brains. Each voxel in the brain template volume was labeled according to ferret brain anatomy and visualized with 3D rendering to show the relative anatomy across cortical, white matter and subcortical structures (figure 5). In the course of optimizing methods for this work, it was observed that the most consequential factors for effective template building were: requirement of isotropic voxel dimensions for both in-vivo and ex-vivo images used to generate the templates, pre-processing correction especially of in-vivo EPI geometric distortions and selection of diffeomorphic and tensor-based registration algorithms along with iterative averaging algorithms for template building.Conclusions
This work has provided a set of ferret brain MRI and DTI templates along with an informed and optimized description of methodological aspects related to acquisition, registration and template building. These tools are to be made available along with a web-based viewer in order to benefit the study of normal and disordered brain anatomy and microstructure in a human-relevant species.Acknowledgements
This work was supported by the CDMRP grants W81XWH-13-2-0018 (SJ) and W81XWH-13-2-0019 (CP) with additional resources from the Centers for Neuroscience and Regenerative Medicine. The authors would like to thank the USUHS Translational Imaging Facility and Alex Korotcov and Asamoah Bosomtwi for in-vivo imaging resources and assistance.References
[1] J.G. Fox, R.P. Marini, Biology and Diseases of the Ferret, Biology and Diseases of the Ferret, (2014). [2] C. Pierpaoli, L. Walker, M. Irfanoglu, A. Barnett, P. Basser, L. Chang, C. Koay, S. Pajevic, G. Rohde, J. Sarlls, TORTOISE: an integrated software package for processing of diffusion MRI data, Book TORTOISE: an Integrated Software Package for Processing of Diffusion MRI Data (Editor ed^ eds), 18 (2010) 1597. [3] M.O. Irfanoglu, P. Modi, A. Nayak, E.B. Hutchinson, J. Sarlls, C. Pierpaoli, DR-BUDDI (Diffeomorphic Registration for Blip-Up blip-Down Diffusion Imaging) method for correcting echo planar imaging distortions, NeuroImage, 106 (2015) 284-299. [4] B. Avants, J.C. Gee, Geodesic estimation for large deformation anatomical shape averaging and interpolation, NeuroImage, 23 Suppl 1 (2004) 50. [5] M.O. Irfanoglu, A. Nayak, J. Jenkins, E.B. Hutchinson, N. Sadeghi, C.P. Thomas, C. Pierpaoli, DR-TAMAS: Diffeomorphic Registration for Tensor Accurate Alignment of Anatomical Structures, NeuroImage, 132 (2016) 439-454.Figures
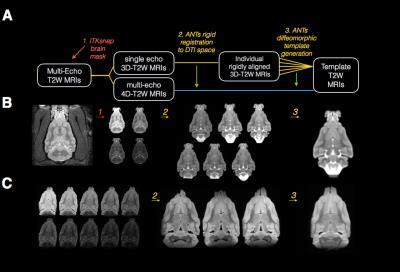
Processing pipeline and template generation for
T2 images of the in-vivo and ex-vivo ferret brain is shown in (A) where rounded
boxes describe the T2 image(s) at each step, numbered italicized text indicates
the software and processing implemented for each step and the same-colored lines
connect the input and output images for each step. Representative in-vivo (B) and ex-vivo (C)
images are shown for each processing step described in (A).
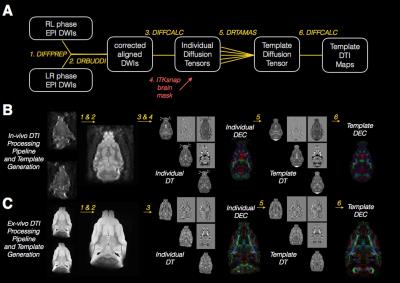
The diagram drawing (A) explains each step of
the DTI processing and template building pipeline where rounded boxes describe
the DWI images and diffusion tensor images at each step, numbered italicized
text indicates the software and processing implemented for each step and the
same-colored lines indicate connect the input and output images for each
step. The red arrow indicates the
indirect use of an ROI from brain segmentation to mask the diffusion tensor
volumes prior to template generation.
Representative in-vivo (B) and ex-vivo (C) images are shown for each
processing step described in (A).
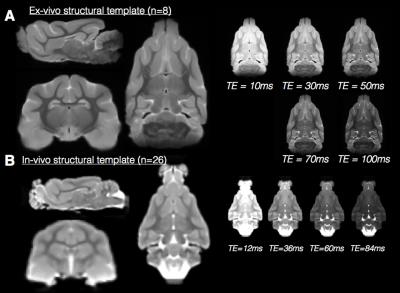
Multiple contrast structural templates in for
in-vivo and ex-vivo ferret brain MRI. Orthographic views (left) of the in-vivo
(A) and ex-vivo (B) structural templates demonstrate the resolution of
neuroanatomical structures for these templates.
A single slice from additional templates generated from the same
multi-echo MRI acquisition are shown to demonstrate the range of tissue
contrasts available (right).
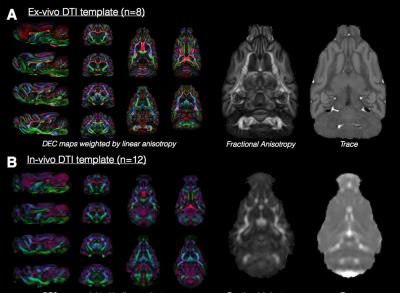
Ex-vivo and in-vivo template DTI maps of the
ferret brain. Representative slices are
shown for the DEC map and a single slice at the same level is shown for Trace
and fractional anisotropy (FA) from ex-vivo (A) and in-vivo (B) DTI. The DTI maps to demonstrate the relative
resolution, smoothing and contrast features of each template DTI map and
between in-vivo and ex-vivo images. The value of n indicates the number of
brain volumes used to generate each template.

Cortical, white matter and subcortical ROI masks
in template space are shown as 3D rendered volumes in the superior, inferior
and oblique views of the brain. Each
region is color-coded according to the legend.