1741
Deconvolution based approaches for the simultaneous quantification of IVIM, Free Water and non-Gaussian behavior in Diffusion MRI.1Department of Information Engineering, University of Padova, Padova, Italy, 2Neuroimaging Lab, Scientific Institute IRCCS Eugenio Medea, Bosisio Parini (LC), Italy, 3Radiology Department, University Medical Center Utrecht, Utrecht, Netherlands
Synopsis
The in-vivo Diffusion MRI (dMRI) signal does not generally arise from a single diffusion process but from the sum of multiples. In this study we investigated a deconvolution approach to simultaneously estimate non-Gaussian diffusion, free water (FW), IVIM and tissue fractions. We analyzed the brain data of a subject acquired with 60 different b-values with four deconvolution based approaches, and compared their results in terms of identified components. The four approaches provided consistent results, with the IVIM compartment being the most similar component across the four methods. Reliable quantification of multiple compartments, including membrane restrictions, is feasible with regularized approaches.
Purpose
The in-vivo Diffusion MRI (dMRI) signal does not arise from a single diffusion process but instead from the sum of multiple processes. Multi-compartmental modeling requires the knowledge of the compartments and is difficult when the number of exponentials is above 2. In this study we investigate a deconvolution approach to simultaneously estimate non-Gaussian diffusion, free water (FW), IVIM and tissue fractions..Methods
The brain data of a healthy subject was acquired with a 3T scanner. The MRI session included a 1mm3 isotropic T1W scan, and a dMRI sequence with multiple b-values (resolution 2.5x2.5x2.5mm3). In particular, dMRI included 3 directions for 20 b-values evenly spaced in the b=0-200s/mm2 range, 20 b-values evenly spaced in the range b=0-1000s/mm2 and 20 b-values evenly spaced in the range b=0-2500s/mm2. Pre-processing of the data included b0 drift removal[1] and affine registration of the volumes to the first b=0s/mm2[2]. A deconvolution dictionary was built as collection of 300 Gaussian decaying diffusion signals with log-spaced diffusion coefficients in the range 0.1-1000.0μm2/ms. An additional column corresponding to zero-diffusion was added to the dictionary to account for non-Gaussian diffusion, i.e. to model the plateu of the diffusion signals after restriction effects. The operation was performed with 4 approaches: (1) sparsity promoted non-negative least squares (NNLS), (2) L2 regularized NNLS[3] (3), L2 regularized NNLS with data estimated priors and (4) L2 regularized NNLS with user-imposed prior. Priors for method (3) were computed first averaging the whole brain in a single voxel, to enhance SNR, then computing the whole brain diffusion spectra with NNLS. Priors for step (4) were imposed by assigning non-zero probabilities to diffusion coefficients that can be found in brain dMRI, as 0.6μm2/ms (slow diffusion), 2.0μm2/ms (fast diffusion), 3.0μm2/ms (FW), 10.0μm2/ms (micro-vascular network) and 200.0μm2/ms (vascular network). Fractional maps were computed by integration of the diffusion spectra in specific diffusion ranges: 6.0-1000.0μm2/ms for the IVIM map, 2.5-6.0μm2/ms for the FW map, 0-2.5μm2/ms for the Tissue Map (TM), and 0-0.4μm2/ms for the slow tissue map (STM). T1W data was segmented with FSL[4], [5] to derive masks of gray matter (GM), white matter (WM) and cerebro-spinal fluid (CSF). Descriptive statistics of the fractional maps were computed to characterize the fractional maps in the three tissues.Results
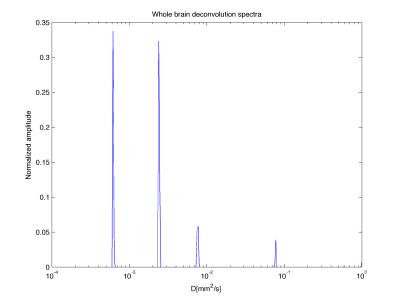
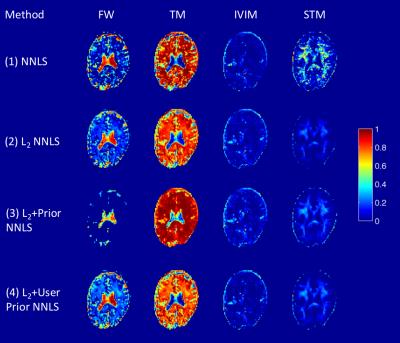
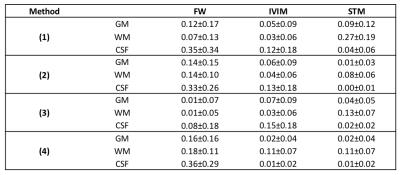
Figure 1 shows the whole brain deconvolution spectra, that was also used as prior for method (3). 5 compartments, corresponding respectively to diffusion coefficients 0.6, 2.4, 7.7, 77.6 and 0μm2/ms (not shown in figure), were revealed. The normalized amplitudes of the peaks were respectively 0.34, 0.32, 0.05, 0.06 and 0.05, therefore the average pseudo-diffusion component was around 10%. Figure 2 shows the fractional maps obtained from the 4 aforementioned methods. Method (1) resulted in the noisiest maps, in particular for STM, while maps from methods (2), (3) and (4) had similar smoothness. FW maps of methods (2) and (4) were similar, and showed higher values than method (3), according to which FW should be zero for most voxels. IVIM maps were consistent across the 4 models. Finally, the STM maps computed with methods (2), (3) and (4) had non-zero values mainly in WM. The mean ± standard deviation of FW, IVIM and STM computed in GM, WM and CSF are reported in Table 1.Discussion
The four approaches provided consistent results. The IVIM compartment was the most consistent across the four methods, with estimated IVIM between 2 and 13%, and higher values in CSF. The STM maps assumed higher values in WM, in agreement with the design of the dictionary to describe diffusion restrictions The deconvolution spectra computed on the whole volume had 5 peaks (Figure 1), however, the peak corresponding to FW was missing. The method might have failed to disentangle FW from fast diffusion tissue and resulted in an average peak. Even if the method is promising, some limitations should be noted. The choice of the priors is critical, as proved by the comparison of methods (3) and (4). A standardized approach to define the priors should be investigated, as well as the choice of the integration ranges to derive fractional maps.Conclusions
Regularized deconvolution with priors can be employed to quickly estimate multiple contributions to the measured diffusion signal. Moreover, the method can be used to quickly remove undesired components before quantification with existing tools.Acknowledgements
No acknowledgement found.References
[1] S. B. Vos, C. M. W. Tax, P. R. Luijten, S. Ourselin, A. Leemans, and M. Froeling, “The importance of correcting for signal drift in diffusion MRI.,” Magn. Reson. Med., vol. 22, p. 4460, Jan. 2016.
[2] S. Klein, M. Staring, K. Murphy, M. A. Viergever, and J. P. W. Pluim, “elastix: a toolbox for intensity-based medical image registration.,” IEEE Trans. Med. Imaging, vol. 29, no. 1, pp. 196–205, Jan. 2010.
[3] B. Madler, D. R. Hadizadeh, and J. Gieseke, “Assessment of a Continuous Multi-Compartmental Intra-Voxel Incoherent Motion ( IVIM ) Model for the Human Brain,” in International Society for Magnetic Resonance in Medicine, 2013.
[4] S. M. Smith, “Fast robust automated brain extraction.,” Hum. Brain Mapp., vol. 17, no. 3, pp. 143–55, Nov. 2002.
[5] Y. Zhang, M. Brady, and S. Smith, “Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm.,” IEEE Trans. Med. Imaging, vol. 20, no. 1, pp. 45–57, Jan. 2001.
Figures