1739
The Maastricht Diffusion Toolbox (MDT): Modular, GPU accelerated, dMRI microstructure modeling1Maastricht University, Maastricht, Netherlands, 2Brain Innovation, Maastricht, Netherlands
Synopsis
MDT's object oriented modular design allows arbitrary user specification and combinations of dMRI compartment models, diffusion microstructure models, likelihood functions and optimization algorithms. Many diffusion microstructure models are included, and new models can be added simply by adding Python script files. GPU based computations allow for ~60x faster model fitting; e.g. the 81 volume example NODDI dataset can be fitted whole brain in about 40 seconds, which makes MDT ideal for population studies. MDT can be extended to other modalities and models such as quantitative MRI. The software is open source and freely available at https://github.com/cbclab.
Introduction
Recent
advances in diffusion MRI (dMRI) modeling propose multi-compartment
microstructure models that promise greater specificity over DTI1,2
and offer additional microstructure measures such as axonal density,
dispersion and diameter distributions. Models of this type include
(but not limited to)
CHARMED3,
NODDI4,
AxCaliber5,
ActiveAx6 and variations7–10.
These multi-compartment models commonly require non-linear model
fitting, which has considerable challenges in quality of fit (local
minima, convergence) and runtime, where runtime in particular is
often prohibitive for larger studies (>20 subjects) and population
studies (>100 subjects). In this work we present a new unified
dMRI microstructure analysis toolbox, the Maastricht Diffusion
Toolbox (MDT),
which
can implement any
microstructure model, comes
pre-supplied with a library of models, has
a unified implementation of cascading (chained optimization) and uses
graphic card accelerated computations that allow for ~60x faster
model fitting over traditional CPU based implementations. The
software is open source and
freely available at
https://github.com/cbclab
under an L-GPL license.Methods
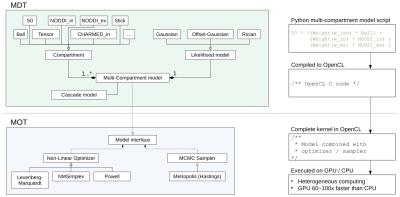
Figure
1 shows a (high level) overview of MDT and the Maastricht
Optimization Toolbox (MOT). In MDT, compartments are combined using a
high level Python-based scripting-language (see upper right in Figure
1) to form, together with a likelihood model, a multi-compartment
model. MDT compiles these models to OpenCL C and uses MOT to optimize
or sample them on the CPU and/or GPU (Graphics Processing Unit;
graphics card) using the OpenCL framework, a free and open standard
for heterogeneous parallelized computations. MDT features cascade
models which offer an unified approach to chained optimization
strategies in which parameters of a complex model are initialized
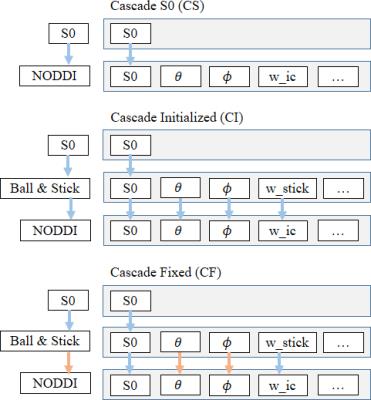
with, or fixed to, values of a simpler model. Figure 2 shows three
cascading strategies used as an example in this work. In the first,
Cascade S0 (CS), the final desired model is initialized only with an
S0 (b0 signal) estimate. Cascade Initialize (CI) extends this by
initializing parameters of complex models from those of simpler
models. Cascade Fixed (CF) is an adaption of CI in which some
parameters are fixed instead of initialized, reducing dimensionality
of later optimization steps. Out of the box, MDT comes with many
popular diffusion models such as Tensor, Ball&Stick, NODDI,
CHARMED and ActiveAx. New models can be added simply by adding Python
script files (see Figure 1 top right). All models can be modularly
combined with any likelihood model and any optimization and sampling
routine. Additional MDT features are: support for provenance trace,
batch fitting routines, detailed logging, data consistency checks and
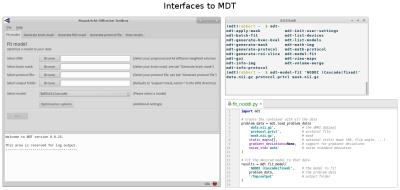
extensibility for optimization and sampling routines. MDT has a
Python interface, a command line interface and a graphical interface
(Figure 3) and is compatible with Linux, Mac and Windows and with
most graphics card and CPU hardware. Processing reported here was
done on a single AMD Fury X graphics card.Results and discussion
To
illustrate the performance of MDT and MOT, we fitted two common
models, CHARMED_in3 (with 3 intra-axonal compartments) and NODDI to
two diffusion MRI datasets of the HCP MGH Consortium1.
Both datasets (1003 and 1004 from the HCP MGH Consortium) were
acquired at a resolution of 1.5mm isotropic with 4 shells of b=1000,
3000, 5000, 10,000, s/mm^2, with respectively 64, 64, 128, 393
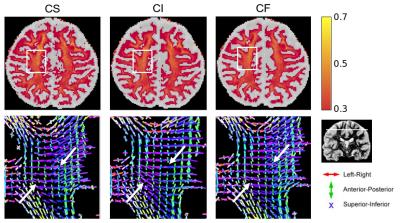
directions and with 40 b0 volumes. Figure 4 shows the fitting results
of CHARMED_in3, highlighting some differences between the three
cascading strategies CS, CI and CF with the Powell optimization
algorithm and the Ball&Stick model as the cascading basis. Whole
brain computation time was ~2 hours for CS, ~2 hours for CI and ~1
hour for CF for this complex model on this 552 volume dataset. Figure
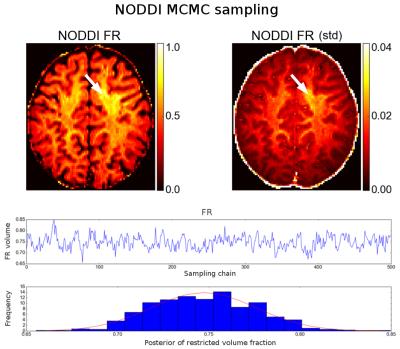
5 shows the results of MCMC sampling (NODDI model fit using CF and
Powell to initializing a random-walk metropolis MCMC sampler, 500
samples of the FR parameter posterior). Fitting and sampling this
single slice took ~12 minutes on the 552 volume dataset. For
smaller and better known datasets, such as the 81
volume example NODDI
dataset, whole brain NODDI can be computed in ~40 seconds.Conclusion
We here put forward MDT, a modular, GPU accelerated, dMRI microstructure modeling and analysis toolbox. The low costs of suitable graphics card (<400 euro) and the ensuing much reduced runtimes as well as the ease of modelling make MDT suitable for rapid model prototyping and whole brain analysis at a single workstation. On a moderate sized server or workstation it is feasible to analyse population studies of hunderds of subjects (see Harms et al. this conference). Finally, by splitting the software stack into MDT and MOT we envision software extensions to other modalities such as light microscopy or quantitative MRI.Acknowledgements
No acknowledgement found.References
1. Assaf Y, Freidlin RZ, Rohde GK, Basser PJ. New modeling and experimental framework to characterize hindered and restricted water diffusion in brain white matter. Magn Reson Med. 2004;52(5):965-978. doi:10.1002/mrm.20274.
2. De Santis S, Drakesmith M, Bells S, Assaf Y, Jones DK. Why diffusion tensor MRI does well only some of the time: Variance and covariance of white matter tissue microstructure attributes in the living human brain. Neuroimage. 2014;89:35-44. doi:10.1016/j.neuroimage.2013.12.003.
3. Assaf Y, Basser PJ. Composite hindered and restricted model of diffusion (CHARMED) MR imaging of the human brain. Neuroimage. 2005;27(1):48-58. doi:10.1016/j.neuroimage.2005.03.042.
4. Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: Practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage. 2012;61(4):1000-1016. doi:10.1016/j.neuroimage.2012.03.072.
5. Assaf Y, Pasternak O. Diffusion tensor imaging (DTI)-based white matter mapping in brain research: A review. J Mol Neurosci. 2008;34(1):51-61. doi:10.1007/s12031-007-0029-0.
6. Alexander DC, Hubbard PL, Hall MG, et al. Orientationally invariant indices of axon diameter and density from diffusion MRI. Neuroimage. 2010;52(4):1374-1389. doi:10.1016/j.neuroimage.2010.05.043.
7. Jelescu IO, Veraart J, Adisetiyo V, Milla SS, Novikov DS, Fieremans E. One diffusion acquisition and different white matter models: How does microstructure change in human early development based on WMTI and NODDI? Neuroimage. 2015;107:242-256. doi:10.1016/j.neuroimage.2014.12.009.
8. Tariq M, Schneider T, Alexander DC, Gandini Wheeler-Kingshott CA, Zhang H. Bingham-NODDI: Mapping anisotropic orientation dispersion of neurites using diffusion MRI. Neuroimage. 2016;133:207-223. doi:10.1016/j.neuroimage.2016.01.046.
9. De Santis S, Jones DK, Roebroeck A. Including diffusion time dependence in the extra-axonal space improves in vivo estimates of axonal diameter and density in human white matter. Neuroimage. 2016;130:91-103. doi:10.1016/j.neuroimage.2016.01.047.
10. Santis S De, Assaf Y, Jeurissen B, Jones DK, Roebroeck A. T1 relaxometry of crossing fibres in the human brain. Neuroimage. 2016;141:133-142. doi:10.1016/j.neuroimage.2016.07.037.
Figures