1724
Interrogating neurotransmitter systems using resting-state fMRIJoanes Grandjean1, Michaela Bürge2, David Bühlmann3, Hannes Sigrist2, Erich Seifritz2, Franz X. Vollenweider2, Christopher R. Pryce2, and Markus Rudin3,4
1Singapore BioImaging Consortium, Agency for Science, Research and Technology, Singapore, Singapore, 2Psychiatric Hospital, University of Zurich, Switzerland, 3Institue for Biomedical Engineering, University and ETH Zürich, Switzerland, 4Institute of Pharmacology and Toxicology, University of Zurich, Switzerland
Synopsis
We assessed functional connectivity (FC) in the mouse brain following psilocybin administration. Conventional analysis using dual regression and reference spatial maps derived from independent component decomposition identified a decrease in FC within the ventral striatum network in animals administered with psilocybin relative to vehicle. Dopamine D1 and serotonin 5HT2a receptor gene expression as well as projection maps from the ventral tegmental area and dorsal raphe nuclei obtained from the Allen Brain Institute database were used as spatial references in a secondary dual regression analysis to disentangle the contribution of dopamine and serotonin systems to whole-brain functional connectivity changes.
Introduction
Functional connectivity (FC) estimated with resting-state fMRI offers a unique window into the brain for characterizing the effects of pharmacological interventions. Many pharmacological agents used to treat psychiatric disorders, such as antidepressants, affect synaptic activity at or binding to receptors related to neurotransmitter systems projecting from midbrain nuclei such as the serotonergic system in the dorsal raphe nucleus (DRN) and dopaminergic systems in the ventral tegmental area (VTA). This is the case for the hallucinogenic drug psilocybin, a 5-HT2A agonist with potential antidepressant effects. Yet, assessing the contribution of individual neurotransmitter systems to resting-state networks (RSNs) remains challenging. RSNs generally do not include midbrain nuclei, and placing seeds on these regions fails to capture functionally coupled distal regions. In this study, we propose a novel approach for assessing the functional connectivity changes induced by psilocybin injection in mice by assessing contributions of individual neurotransmitter systems to resting-state signal using gene expression and projection fields within the dual-regression framework.Method
Mice were anesthetized using medetomidine / isoflurane (Grandjean et al. 2014). Psilocybin (1mg/kg n=13 or 2mg/kg n=12) or vehicle (n=15) was injected i.v. 20 min before acquisition of mutli-echo EPI scans (800 volumes, TR=1500ms, TE=[11,17,24]ms) at 9.4T with a 2x2 cryoprobe. Images were processed with meica.py script (Kundu et al. 2012). Standard analysis included dual-regression (Filippini et al. 2009) using 17 reference RSNs identified previously (Zerbi et al. 2015). Structural connectivity maps for midbrain nuclei derived from anterograde viral tracer injection in the DRN and VTA, as well as gene expression maps for serotonin 5HT2a and dopamine D1 receptors, were obtained from the Allen Brain Institute database (http://mouse.brain-map.org/) and transformed to reference space. Injection sites and white matter fibers were masked from tracer injection maps. Tracer and gene expression maps were thresholded to the 90th percentile and used as spatial reference maps in dual-regression analysis. Statistical analysis comparing psilocybin 1mg/kg or 2mg/kg to vehicle, and combined psilocybin effect (1 and 2 mg/kg) to vehicle was carried using nonparametric permutation testing with threshold-free cluster enhancement inference with Bonferroni correction.Results
Dual-regression analysis identified robust psilocybin effects relative to vehicle in the ventral striatum network, indicating decreased FC in psilocybin treated animals (Figure 1). The highest significant cluster from this analysis was used as a reference in seed-based analysis (Figure 2): one-sample t-tests carried out on the seed-based maps revealed FC relative to a seed in the ventral striatum that were confined to the striatum in the vehicle group, whereas FC extended to prefrontal cortex and midbrain in the two psilocybin groups. The seed coordinate was converted to Allen atlas space and used to determine gene expression clusters using the Anatomic Gene Expression Atlas (AGEA). The AGEA cluster revealed spatial distribution comparable to the seed-based map. In particular, both dopaminergic D1 and D2 receptors were expressed in this cluster (1.75 and 2.06 fold change in expression in the AGEA cluster relative to the remaining voxels, respectively). To further investigate the role of dopamine and serotonin on the resting-state signal, in situ hybridization maps for D1 and 5HT2a receptor genes were used as spatial reference maps in dual-regression analysis (Figure 3). With respect to D1 gene expressing regions, psilocybin decreased FC within the striatum, prefrontal cortex, hippocampus and midbrain. With respect to 5HT2a gene expression map, psilocybin increased FC in the striatum. Comparable results were obtained when the viral tracer maps from the VTA and DRN were used as spatial references in the dual regression analysis (Figure 4).Discussion
Gene expression and structural connectivity maps related to the dopaminergic and serotonergic system were used as spatial reference maps in dual-regression analysis in order to discriminate two opposing effects of psilocybin on FC, a) decreased in FC within the ventral striatum related to dopaminergic activity and b) increased striatum coupling to 5HT2a receptor expressing regions and projection field from the DRN. Combining FC analysis with online databases offers a new perspective to gain insights into whole-brain mechanisms of pharmacological agents and to capture different dynamics in neurotransmitter systems.Acknowledgements
No acknowledgement found.References
Filippini, N., MacIntosh, B.J., Hough, M.G., Goodwin, G.M., Frisoni, G.B., Smith, S.M., Matthews, P.M., Beckmann, C.F. and Mackay, C.E. 2009. Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc Natl Acad Sci U S A 106(17), pp. 7209–7214. Grandjean, J., Schroeter, A., Batata, I. and Rudin, M. 2014. Optimization of anesthesia protocol for resting-state fMRI in mice based on differential effects of anesthetics on functional connectivity patterns. Neuroimage 102 Pt 2, pp. 838–847. Kundu, P., Inati, S.J., Evans, J.W., Luh, W.-M. and Bandettini, P.A. 2012. Differentiating BOLD and non-BOLD signals in fMRI time series using multi-echo EPI. Neuroimage 60(3), pp. 1759–1770. Zerbi, V., Grandjean, J., Rudin, M. and Wenderoth, N. 2015. Mapping the mouse brain with rs-fMRI: An optimized pipeline for functional network identification. Neuroimage 123, pp. 11–21.Figures
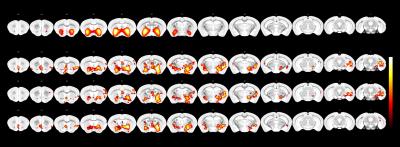
Reference map for the ventral striatum RSN and voxel-wise statistical
maps estimated with dual-regression indicate decreased FC within the RSN in the
psilocybin group (combined 1 and 2 mg/kg groups, 1mg/kg, 2mg/kg) relative to
vehicle.
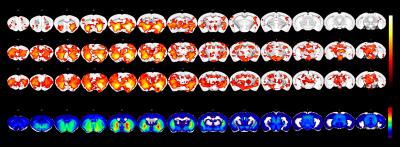
One-sample t-test
maps estimated for seed-based maps estimated with a 2x2 voxel seed region
placed in the ventral striatum. FC is confined to the ventral striatum in the
vehicle group, while it extends to the prefrontal cortex and midbrain in both
psilocybin groups. The Anatomic Gene Expression Atlas cluster for the
seed-region indicates a gene expression related cluster displaying comparable
spatial extent to that defined with FC in the vehicle group. Among the genes
presenting enhanced expression within the cluster are D1 and D2 dopamine
receptors.
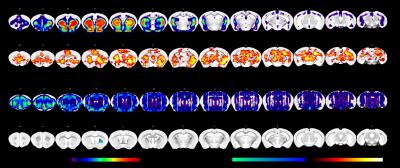
D1 and 5HT2a gene
expression maps were obtained from the Allen Brain Institute used as spatial
references in a dual-regression analysis. Statistical maps indicating
psilocybin effects (combined 1 and 2 mg/kg groups) relative to vehicle in the
striatum and prefrontal and anterior cingulate cortex with respect to D1
receptor expression. With respect to 5HT2a receptor gene expression, FC to the
striatum was increased following psilocybin relative to vehicle.
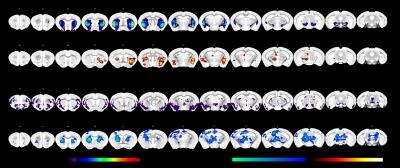
VTA and DRN viral
tracer structural connectivity maps were obtained from the Allen Brain
Institute used as spatial references in a dual-regression analysis. Statistical
maps indicating psilocybin effects (combined 1 and 2 mg/kg groups) relative to
vehicle in the striatum with respect to VTA projection fields. With respect to
DRN projection fields, FC to the striatum, thalamus and midbrain was increased
following psilocybin relative to vehicle.