1650
Correction of EPI geometric distortion in slice direction using reversed slice-select gradients and topup1A.A. Martinos Center for Biomedical Imaging, MGH/Harvard, Charlestown, MA, United States, 2Division of Health Sciences and Technology, Harvard-MIT, Cambridge, MA, United States
Synopsis
Conventional single-shot Echo Planar Imaging (EPI) protocols used by fMRI studies suffer from geometric distortion occurring in the phase-encoding direction. Ultrahigh field (>3T), high-resolution fMRI using thin slices requires decrease of the slice-encoding bandwidth, which may lead to the additional geometric distortions and “slice bending” in the slice-encoding direction. Here we use gradient-echo (GRE) EPI sequence that allows the polarity of the slice-select gradient to be flipped between positive and negative values to demonstrate this effect and to enable the application of FSL’s topup to perform distortion correction. This correction will further improve geometric alignment of the EPI data with anatomical reference data used in fMRI studies.
Introduction
Conventional single-shot Echo Planar Imaging (EPI) protocols widely used by fMRI studies suffer from geometric distortion occurring in the phase encoding direction, which originates from the local magnetic field offsets and is caused by the low phase-encoding bandwidth of the acquisitions (such as 16 Hz/pixel) compared to much higher readout bandwidth (such as 1000 Hz/pixel) (1). Ultrahigh field (>3T), high resolution fMRI using thin slices requires decrease of the slice encoding bandwidth, which may lead to the additional geometric distortions and “slice bending” in the slice-encoding direction. Similar to other distortions this causes problems when EPI acquisitions are aligned to other scans such as MPRAGE. In this work we demonstrate the effect of low slice-select pulse bandwidth on geometric distortion, i.e., local slice shifting, using acquisition allowing reversed polarity of the slice-encoding gradient (a method we call over-easy). In addition, we use data acquired with slice gradient of opposite polarities to perform distortion correction in the slice-encoding direction using the approach implemented in FSL’s topup (2–4), in order to allow alignment of the EPI data with other relatively undistorted structural data often used as an anatomical reference in fMRI studies.Background
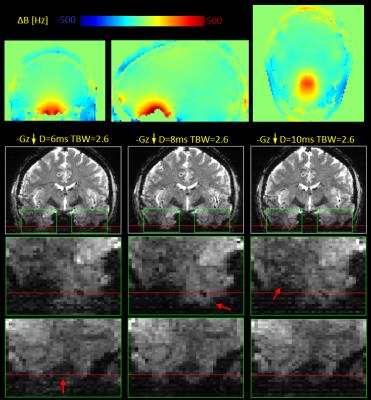
Slice thickness (TH) of the MRI volume is determined by slice encoding gradient strength (Gz) and bandwidth (BW): TH=BW/γGz. While Gz is usually determined by the desired slice thickness, the RF pulse BW can be modified by changing pulse duration (D) or the time-BW product (TBW): BW=TBW/D. It often occurs in sub-millimeter imaging that the slice-select gradient cannot achieve a desired slice thickness for a desired RF pulse characteristic. Thus slice thickness can be decreased by modifying the RF pulse design either by reducing TBW (which leads degradation of the slice profile), or by increasing pulse duration (which lowers SAR but increases minimal TE available for the acquisition). For example, to excite a slice of TH=1mm using a typical slice gradient strength available in 7T, BW in range of 300-800Hz would be required. Therefore, the local magnetic field offsets seen at 7T, which can reach values of 500 Hz (see Figure 1), can cause local shifts of 1-2 slices.Methods
Two healthy volunteers (1F/1M, 23/29yo) were scanned on a whole-body 7T scanner (Siemens Healthcare, Erlangen, Germany) using gradient-echo (GRE) EPI protocol allowing flipping polarity of the slice encoding gradient between positive and negative, with appropriate shifting of the RF pulse center frequency. Two sets of scans using different slice orientation were acquired in resolution 1.5 x 1.5 mm and slice thickness of 1 mm for axial and 1.5 mm for coronal data. Different RF pulse lengths and different time-bandwidth product values were used to control the slice-select BW, as summarized in Table 1. Sets of 10 volumes, 5 for each polarity of the slice encoding gradient, were concatenated and distortion corrected using FSL’s topup. In order to compare slice shifts occurring in both distortion directions, the images were co-registered using Robust Register (5).Results
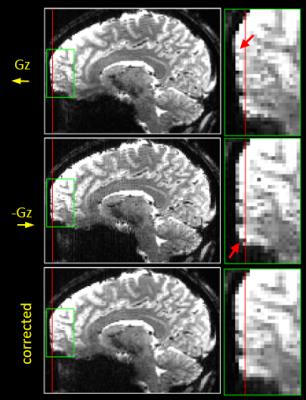
An example B0 field map showing local magnetic field offsets occurring at 7T is presented in Figure 1 together with the example slice shifts caused by these offsets. Figures 2 and 3 present example reformats of the original distorted data acquired with positive and negative polarity of the slice-encoding gradient and the result of topup distortion correction, for data sets acquired natively in both axial and coronal planes.Discussion
The results showed geometric distortions in the EPI data due to the reduced bandwidth in the slice-encoding direction. Here GRE EPI acquisitions were used to acquire the data with flipped polarity of the slice gradient, and future work will be to implement this within a spin-echo acquisition to refocus through-plane dephasing, which is more challenging due to the use of slice-selective excitation and refocusing pulses. The data acquired with opposite slice gradient encoding directions allowed distortion correction with using topup, but this slice-select gradient reversal can also be used as a means to validate geometric distortion correction based on acquired B0 field maps. Gradient-reversal-based distortion correction methods such as topup are beneficial in that it reduces scanning time (only two additional frames are required) and avoiding co-registration challenges due to non-matching distortion and resolution between the field map and EPI data. It may also be possible to combine slice-select gradient reversal with phase-encoding gradient reversal to acquire two volumes with opposite distortions in both directions to correct for both forms of distortion simultaneously, further reducing the required scan time needed for mapping these distortions.Conclusions
Acknowledgements
Supported by NIH NIBIB P41-EB015896 and R01-EB019437, NIMH R01-MH111419 (BRAIN Initiative), the Athinoula A. Martinos Center for Biomedical Imaging, and made possible by NIH NCRR Shared Instrumentation Grants S10-RR023401, S10-RR023043 and S10-RR020948.References
1. Jezzard P. Correction of geometric distortion in fMRI data. NeuroImage 2012;62:648–51.
2. Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage 2004;23 Suppl 1:S208–19.
3. Andersson JLR, Skare S, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. NeuroImage 2003;20:870–88.
4. Holland D, Kuperman JM, Dale AM. Efficient Correction of Inhomogeneous Static Magnetic Field- Induced Distortion in Echo Planar Imaging. NeuroImage 2010;50:1–18.
5. Reuter M, Rosas HD, Fischl B. Highly accurate inverse consistent registration: A robust approach. NeuroImage 2010;53:1181–1196.
Figures