1587
Segmentation of CMF Bones from MRI with A Cascade Deep Learning Framework1Department of Radiology and BRIC, UNC-Chapel Hill, USA, Chapel Hill, NC, United States, 2Houston Methodist Hospital, Houston, TX, USA
Synopsis
Purpose
Magnetic Resonance Imaging (MRI) provides a safe and non-invasive way to study internal tissues and create detailed Craniomaxillofacial (CMF) surgical reference model for planning. However, obtaining these models requires manually segmenting bones, which is a prohibitively time-consuming process (segmenting one brain alone takes an expert several hours). Automatic segmentation could allow the mass generation of bone models. In this paper, we propose a cascade deep learning framework to automatically segment CMF bone tissue out of the MR images for brain. Firstly, we train a 3D fully convolutional network (FCN) model for segmentation of MRI brain images in an end-to-end and voxel-to-voxel fashion. Secondly, to acquire better segmentation maps, we propose to use an additional convolutional neural network (CNN) to further classify voxels predicted to be bone tissue from the initial segmented maps. Note, we adopt a batch normalization (BN) strategy to better solve the potential optimization problems, thus making the model converge at a faster speed and also increasing discrimination capability.Method
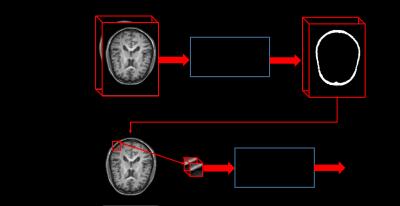
Described by [1], FCN actually consists of two kinds of operations: down-sampling and up-sampling. The down-sampling operation streams (i.e., convolution and pooling) usually result in coarse and global predictions based on the entire input of the network; and the up-sampling streams (i.e., deconvolution) could generate dense prediction through finer inference. We take advantage of Segnet [2] architecture directly, and just adjust the filters to be 3D (denoted as 3D-Segnet). Specifically, we utilize a patch size of 152×184×5 as input for the 3D version of Segnet, by which we seek to acquire the coarse bone tissue structure. We know that higher-layer features induce higher-abstraction of the input, and therefore they can easily ignore small-structure bones in the 3D image. Thus, it usually intends to predict the bones to be thicker. To solve this issue, we further propose to use an additional convolutional neural networks (CNN) [3] (again, we adjust it for 3D operations and set the number of output units to be 2, denoted as 3D-VGG) to further judge if the voxel from the predicted bone region belongs to bone tissue or not. Note, we propose to use small patches 15×15×15 for each voxel in the predicted bone tissue region from the initial segmented maps. In this way, we can refine the initial segmented maps and alleviate the thickness problem casued by FCN. The whole cascade deep learning framework is shown in Fig. 1. In the whole framework, a batch normalization [4] operation is adopted after each convolution operation to make the network easier to converge.
Results
We conduct experiments on a 8-subject dataset with MR images for brain in a leave-one-out fashion.To qualitatively demonstrate the advantage of the proposed method on this dataset, we first show the segmentation results of different tissues for a typical subject in Fig. 2.
To quantitatively evaluate the segmentation performance, we use Dice ratios to measure the overlap between automated and manual segmentation results. Specifically, the proposed intial segmentation (3D-Segnet) method could achieve average Dice ratio of 0.8382±0.0319; with the proposed refine segmentation method (3D-VGG), we could achieve average Dice ratios of 0.9307±0.0350.
Conclusion
In this paper, we have proposed a cascade deep learning based method to segment CMF bones from with MR images of the brain. In our designed segmentation framework, we first propose to use FCN to do an initial segmentation. With these initial segmented maps, we then focus on the predicted bone regions, that is to say, we propose to use CNN to do binary classification for each voxel in the bone region. Our proposed method could achieve good performance in terms of segmentation accuracy. In addition, our proposed segmentation framework could extend to other bone segmentation tasks.Acknowledgements
This project is sponsored in part by NIH/NIDCR DE022676 and DE021863.References
[1] Long, J., E. Shelhamer, and T. Darrell, Fully convolutional networks for semantic segmentation. arXiv preprint arXiv:1411.4038, 2014.
[2] Badrinarayanan, Vijay, Alex Kendall, and Roberto Cipolla. "Segnet: A deep convolutional encoder-decoder architecture for image segmentation." arXiv preprint arXiv:1511.00561 (2015).
[3] Simonyan, Karen, and Andrew Zisserman. "Very deep convolutional networks for large-scale image recognition." arXiv preprint arXiv:1409.1556 (2014).
[4] Ioffe, S. and C. Szegedy, Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:1502.03167, 2015.