1533
Automated Segmentation of Knee Cartilage from MR Images using Sequential Multi-atlas Registration and Shape-constrained Locally-weighted Voting for Quantitative Knee Joint Assessment1School of Electrical Engineering, KAIST, Daejeon, Korea, Republic of, 2Dept. of Software Convergence, Seoul Women's University, Seoul, Korea, Republic of, 3Department of Radiology, Samsung Medical Center, Seoul, Korea, Republic of
Synopsis
We propose a multi-atlas segmentation method for the knee cartilage in T2 PD MR images using sequential multi-atlas registrations and locally-weighted voting (LWV). To select training atlases similar to the test image, a 2D projection image-based atlas selection method is proposed. Then, to extract a bone model to be used as registration target in cartilage segmentation, the bone is segmented by sequential multi-atlas registrations and LWV. Finally, to segment a cartilage without leakage into low-contrast surroundings, the cartilage is segmented by bone-mask-based cartilage registration and shape-constrained LWV with distance and structure similarity weights, as well as atlas similarity weight.
Purpose
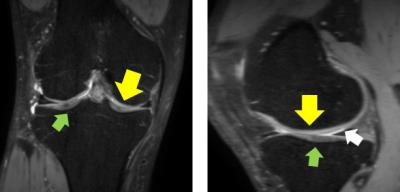
Knee cartilage is a soft tissue in knee joint which plays an important role in maintaining its stability. Quantitative assessment of cartilage morphometry in MR images has been used as an important tool for a number of clinical tasks including diagnosis, disease tracking, and treatment planning of osteoarthritis (OA). However, the segmentation of cartilage is known to be challenging due to (1) its thin plate-like shape and variation in it, (2) the low contrast surroundings, e.g. synovial fluid (Fig. 1), and (3) adjacency between femoral and tibial cartilages. To overcome the challenges, we propose an automated segmentation of cartilage from knee MR images using sequential multi-atlas registration and shape-constrained locally-weighted voting (LWV).Methods
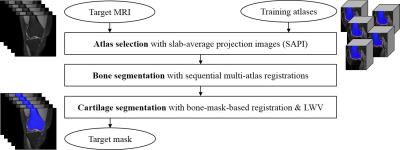
3D T2 PD VISTA coronal knee joint MRI scans were acquired for fourteen normal subjects, using Philips Achieva 3.0T system with a repetition time (TR) of 1600ms, an echo time (TE) of 32.69ms, a resolution of 512x512, a slice number of 250, a pixel size of 0.31mm x 0.31mm, and a slice thickness of 0.5mm. To construct the training atlases, bones and cartilages for all MRI scans were manually segmented by three experts. For each test image, the bone and cartilage were segmented by the proposed method consisting of three main steps (Fig. 2). First, an atlas selection is performed to reduce the computation time and registration error in multi-atlas registration. In atlas selection, the training atlases similar to the test image are selected by registering and comparing slab average projection images (SAPI) instead of 3D scans. Second, the bone was segmented by 2-stage sequential registrations1 and LWV with atlas similarity weight.2 Bone was first roughly segmented by volume-based 3D affine registration and LWV, and then refined by object-based 3D affine registration, and LWV. Third, the cartilage was segmented by bone-mask-based 3D affine registration and shape-constrained LWV. To reduce the registration error and to enhance the robustness to the cartilage shape variability, the cartilage masks of the training images were registered to the test image using 3D affine registration between bone masks. In addition to conventional atlas similarity weight, two additional shape-constrained weights, distance similarity and structure similarity, for LWV were proposed to reduce the leakage into low contrast surroundings in cartilage segmentation. A distance similarity weight weighs the distance between each voxel and bone surface, based on the fact that the cartilage is thin plate stuck on the bone surface. A structure similarity weight emphasizes the structural similarity between the registered training cartilage masks and the test cartilage mask based on structural tensor, especially the measurement of plate-likeliness.3Results
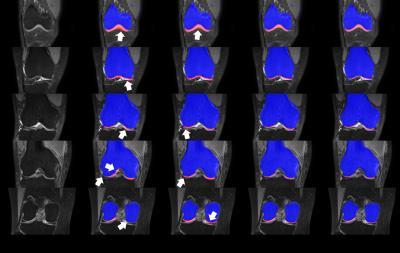
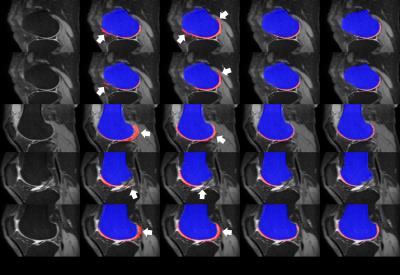
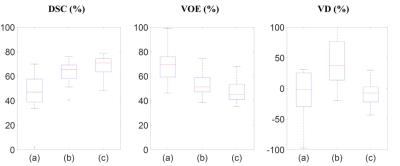
In evaluation, two comparative methods, with (1) the conventional volumetric bone and cartilage registration and globally-weighted voting (GWV), and (2) the sequential registrations and GWV, were compared with the proposed method in qualitative and quantitative assessment. (Figs. 3, 4) Experiments were cross-validated with leave-one-out method. In qualitative assessment, it can be observed that the proposed method reduced misaligned segmentation compared to Volume+GWV with the help of sequential registrations. It can also be observed that the proposed method avoided the leakage into low-contrast surroundings compared to Sequential+GWV, thanks to shape-constrained LWV. In quantitative assessment, the proposed and comparative methods were assessed with the manual segmentation using Dice similarity coefficient (DSC), volume overlap error (VOE), and volume difference (VD),4 as shown in Fig. 5. It can be observed that the proposed method outperformed the comparative methods by achieving the DSC, VOE, and VD of 68.9±8.1%, 46.9±9.0%, and -7.5±19.0%, respectively.Discussion
In our method, the proposed sequential multi-atlas registration reduced the misalignment between the training atlases and the target image without using non-rigid registration techniques. Our shape-constrained LWV enabled our method to avoid the leakage into low contrast surroundings. As a result, our method enabled the segmentation to be robust to the cartilage shape variability, and to prevent the leakage into low contrast surroundings.Conclusion
We have developed an automated cartilage segmentation method in knee MR images using sequential multi-atlas registration and shape-constrained LWV. Our method can be applied to quantitative assessment of cartilage morphometry for various OA-related tasks, including cartilage loss tracking for OA diagnosis and 3D print model estimation for reconstruction surgery.Acknowledgements
Our work was supported by grants of Samsung Electronics Digital Media and Communications R&D center, and the Ministry of Science, ICT & Future Planning (MISP), Korea, under the National Program for Excellence in SW (R7116-16-1018) supervised by the Institute for Information & Communications Technology Promotion (IITP).References
1. Lee H.S., Kim H.A., Kim H., Hong H., Yoon Y.C., Kim J. Multi-atlas segmentation of the cartilage in knee MR images with sequential volume- and bone-mask-based registrations. SPIE Med. Img. 2016.
2. Lee, J-G., Gumus, S., Moon, C.H., Kwoh, C.K., and Bae, K.T. Fully automated segmentation of cartilage from the MR images of knee using a multi-atlas and local structural analysis method. Medical Physics 2014; 41(092303): 1-10.
3. Lee, J.-G., Kim, J.H., Kim, S.H., Park, H.S., and Choi, B.I. A straightforward approach to computer-aided polyp detection using a polyp-specific volumetric feature in CT colonography. Comput. Biol. Med. 2011; 41(9): 790-801.
4. Heimann, T., Morrison, B., Styner, M., Niethammer, M., and Warfield, S. Segmentation of knee images: A grad challenge. Proc. MICCAI Works. Med. Img. Anal. Clinic, 2010.
Figures