1519
A method for identifying and fixing faulty navigator corrections in system-reconstructed multi-shot 3D diffusion weighted images1Department of Animal Science, The Pennsylvania State University, University Park, PA, United States, 2Department of Biomedical Engineering, The Pennsylvania State University, University Park, PA, United States, 3Huck Institutes of the Life Sciences, The Pennsylvania State University, 4Interdepartmental Graduate Program in Neuroscience, The Pennsylvania State University
Synopsis
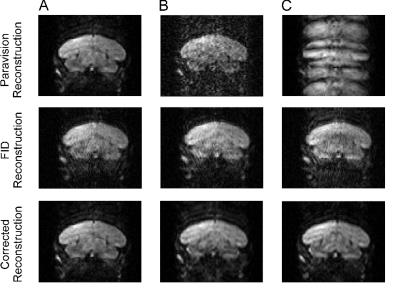
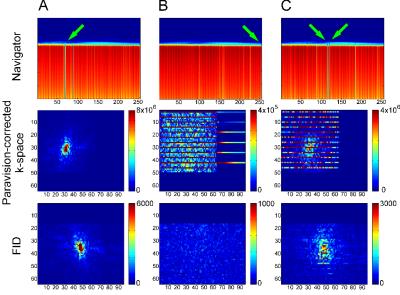
Unusual artifacts appear in some reconstructed images of a multi-segment 3D EPI-DTI sequence with navigator correction on a 7T Bruker Biospec system running Paravision 6.1. Navigator helps to compensate for subject movement within the scanner. However, by examining the k-space of the system processed volumes and the raw navigator data, the artifacts were attributed to navigator overcompensation, which resulted in the over representation of a few lines in k-space. After zeroing the affected lines, image quality was on par with other volumes with no artifacts, eliminating the need to rescan subjects.
Introduction
In echo planar imaging (EPI) subject motion during acquisition can cause a translation or rotation of the points in k-space, which can result in image blur and/or ghosting artifacts1. Navigator echoes interspersed through the scan may be used to correct the distortions in k-space2. In EPI multiple lines of k-space are acquired between radio frequency (RF) excitation pulses, reducing total scan time. In multi-shot EPI diffusion tensor imaging (EPI-DTI), each slice phase is acquired in segments to reduce image distortion due to fast gradient switching and increase the signal to noise ratio (SNR) 3 at the cost of longer scan times.
During 3D multi-shot EPI-DTI, navigator pulses at the beginning of each segment are used to make phase adjustments which correct for subject movement during acquisition. When imaging songbird brains, despite rigid head constraints and anesthesia, small movements are occasionally observed in the respirometer waveform. In the majority of scans, the navigator phase correction functions as it should, resulting in an image with good spatial contrast and low levels of ghosting, even when movement is observed. However, we have observed instances where the correction algorithm overcompensates (in 18 experiments, 2 were affected) resulting in severe artifacts, which would normally require the subjects to be rescanned. The affected volumes showed severe ghosting or high noise (see Figure 1). By looking at the k-space of the reconstructed volumes, we were able to isolate and fix errors in reconstruction. Here we describe the process for determining that the images were corrupted by the navigator and present a simple method for correction allowing the experimenter to use what would otherwise be "throwaway" data.
Image Acquisition
White throated sparrows (Zonotricia albicollis) were imaged on a 7T Bruker Biospec (Billerica, MA) horizontal bore system running Paravision 6.1. Animals were anesthetized initially using 1-2.5% isoflurane in oxygen. The animal’s head was fixed using a custom 3D printed head holder with ear-bars and a beak-bar. The breathing rate was monitored throughout the experiment via a respirometer pressed against the animal’s chest. Despite the anesthesia and head constraint, occasionally the animal kicks while inside the scanner, resulting in small movements during the scan. A Rapid Biomedical (Rimpar, Germany) quadrature birdcage resonator and double surface coil were used respectively for RF transmit and receive. DTI sequences were acquired in two blip directions (TE=20.1ms, TR=1s, Segments=4, resolution=230x340x340µm2, matrix size=96x48(partial-k, zero-filled to 64)x64, averages=1, directions=20, b0 volumes=2, b value=1000, scan time=3h6m).Data Conditioning and Image Processing
Paravision reconstructed images were saved as complex images in 2dseq format. These are converted to nifti format
using Bru2nii, an open-source tool created by Chris Rorden4. Custom
scripts are used to import the raw FID (free induction decay) file and complex nifti file into a Matlab (The Mathworks; Natick,
MA) workspace. A 3D fast Fourier transform (FFT) is applied to the
Paravision-reconstructed volumes. Those volumes that are corrupted by
overzealous navigator correction have lines in k-space that are much higher in
amplitude than the average line. They cover the slice in k-space but only every
fourth line is affected (see Figure 2). Among the navigator segments of the data, the
amplitude is much lower preceding the acquisition of the affected lines in
K-space. We used a custom Matlab script to to zero-fill the lines that were
affected, after identifying sub-threshold navigator traces. Finally, the corrected
image is reconstructed again with an inverse Fourier transform of the k-space.
Discussion
Images that were corrupted by the erroneous navigator correction appeared to be completely unusable. Artifacts included severe ghosts, and image blurring. Ghosts occurred when the navigator error occurred near the middle of k-space. Blurring occurred when navigator error occurred near the edge of k-space. Diffusion tensors calculated with the remaining (unaffected) diffusion weighted images suffer from decreased angular resolution due to using fewer and non-uniformly distributed directions. After zero-filling the affected lines in k-space, the reconstructed volumes were visually normal, and the data could then be entered into the normal processing pipeline.Acknowledgements
This work was supported by the Office of Naval Research Awards N00014-16-1-2187 and N00014-14-1-0703 to Paul Bartell.References
1. Axel, L., et al. (1986). doi:10.1148/radiology.160.3.3737920
2. Ehman, R. L., & Felmlee, J. P. (1989). doi:10.1148/radiology.173.1.2781017
3. Chen, N., & Wyrwicz, A. M. (2004). doi:10.1002/mrm.20097
4. Rorden, C. (2016). Retrieved from https://github.com/neurolabusc
Figures