1500
Accelerated 3D GRASE for T2 and PD Weighted High Resolution Images1Department of Radiology and Nuclear Medicine, Erasmus MC, Rotterdam, Netherlands, 2GE Healthcare B.V., Hoevelaken, Netherlands
Synopsis
Parallel Imaging techniques have not been introduced for Gradient and Spin Echo sequences being a limiting factor for clinical use. Enabling PI for GRASE requires new view-ordering schemes that acquire an autocalibration region while simultaneously mitigating artifacts and obtaining the desired contrast. The purpose of this work is to present new 2D PI accelerated Cartesian view-ordering schemes with either T2 or PD contrast in multi-shot VFA 3D-GRASE, for relevant SAR and scan time reduction compared to 3D-FSE/TSE.
Introduction
Gradient and Spin Echo sequence (GRASE1) has recently been proposed as an alternative to FSE/TSE for structural MRI in high field MRI (7T), where the specific absorption rate (SAR) limits spatial resolution2. GRASE is increasingly used for fMRI3 and Arterial Spin Labelling (ASL)4 since higher time and spatial resolution can be combined with less susceptibility artifacts compared to EPI5.
Currently, a limiting factor for GRASE is that Parallel Imaging techniques (PI)6 have not been developed for GRASE. Enabling PI for GRASE requires new view-ordering schemes that acquire an autocalibration region while simultaneously mitigating artifacts and obtaining the desired contrast. The purpose of this work is to present new 2D PI accelerated Cartesian view-ordering schemes with either T2 or PD contrast in multi-shot VFA 3D-GRASE, for relevant SAR and scan time reduction compared to 3D-FSE/TSE.
Methods
Two view-ordering techniques are combined to enable parallel imaging in 3D-GRASE: SORT7, which splits off-resonance and T2 effects in different phase encoding directions, and linear modulation8 , which mapping the signal modulation into k-space to include 2D acceleration, achieve different image contrasts and mitigate artifacts.
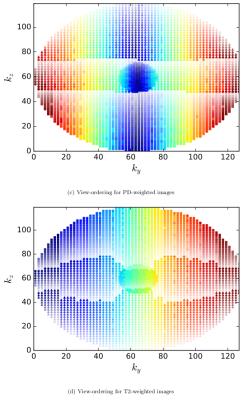
The algorithm uses any k-space sampling pattern, as list of k-space positions, the Echo train length (ETL) and EPI factor (N) as input. To each k-space position it assigns a train number, echo number along the ETL, and the echo in N. To obtain PD-weighted contrast, the center of the k-space has to be filled at the beginning of the Echo Train (ET). This is accomplished by separating the positions with $$$k_{y} \geq k_{y\_center}$$$ and $$$k_{y}<k_{y\_center}$$$ and sorting on $$$k_{z}$$$ each sublist. Subsequently, each sublist is split in N equally sized parts (to make the size equal, the N sublist is completed with zeros), , where list number defines the echo in N. Each sublist is sorted on $$$k_y$$$ and split in ETL parts, specifying echo number, each sorted on $$$k_z$$$. The position in the final list specifies the train number, except that the $$$k_{y} \geq k_{y\_center}$$$ and $$$k_{y}<k_{y\_center}$$$ lists are interleaved along the acquisition (See Figure 1a). To achieve T2-weighted contrast, the center of k-space should be filled at the middle of the ET. For this, the list of k-space positions is sorted on $$$k_{y}$$$ and split in ETL equally sized parts, specifying echo number, sorted on $$$k_{z}$$$. The position in this list divided by N specifies the train number while the position modulo N specifies the echo in N (See Figure 1b) .
Additionally one reference echo train without playing out slice and phase gradients is acquired for phase and amplitude correction9. Echoes were time corrected by shifting the echo peak to the center of the time acquisition window. Phase correction was performed by point by point subtraction of the phase of the Fourier transformed reference echoes from the Fourier transformed echoes in the acquisition. Subsequently, the images are reconstructed with Autocalibrating Reconstruction for Cartesian sampling (ARC)10,11, with sum of squares channels combination. Finally, a correction for gradient non-linearities was applied to each slice.
Results
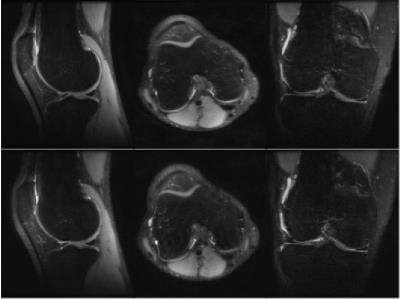
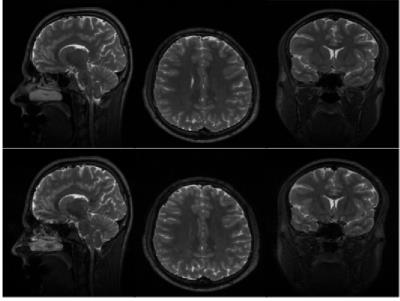
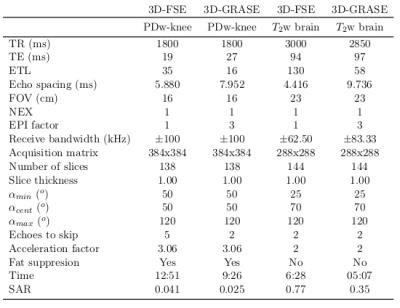
Human in vivo experiments were performed on a 3T GE MR750 Scanner with a four channel transmit-receive knee coil for whole knee PD-weighted images and a 8-channel receive head coil for whole brain T2-weighted images. Table 1 shows the acquisition parameters, time and the SAR values for VFA 3D-GRASE and VFA 3D-FSE in a knee and a brain. The parameters used for the GRASE acquisition were chosen to maintain the same TEeff as FSE. Figure 2 and 3 shows three orthogonal slices of the PD-weighted images of a knee and the T2-weighted images of a brain, respectively, of VFA 3D-FSE/TSE and VFA 3D-GRASE for the view-orders proposedDiscussion
A high reduction in acquisition time (~25%) and SAR (~50%) is obtained with VFA PI 3D-GRASE compared to VFA PI 3D-FSE. The three orthogonal slices visually show no artifacts or distortions. Higher bandwidth was used for 3D-GRASE to maintain the ESP as short as possible, in order to achieve short TE and to avoid off-resonance effects between refocusing pulses, although it can increase the noise level in the images.Conclusion
The presented view-ordering schemes for VFA 3D-GRASE have demonstrated to enable PI acquisitions for T2 and PD contrast without apparently loss of contrast and resolution with a relevant reduction on the scan time compared to 3D-FSE/TSE. These flexible view-order schemes are easily extended for Compressed Sensing, achieving a higher reduction in SAR, compared to FSE.Acknowledgements
This work was supported by a GE grant. We would like to thank Rob Peters, Hendrik de Leeuw, Gyula Kotek and Piotr Wielopolski.References
[1] Koichi Oshio and David A Feinberg. GRASE (gradient-and spin-echo) imaging: A novel fast MRI technique. Magnetic Resonance in Medicine, 20(2):344–349, 1991.
[2] Robert Trampel, Enrico Reimer, Laurentius Huber, Dimo Ivanov, Robin M. Heide- mann, Andreas Schäfer, and Robert Turner. Anatomical brain imaging at 7T using two-dimensional GRASE. Magnetic Resonance in Medicine, 72(5):1291–1301, 2014.
[3] Valentin G. Kemper, Federico De Martino, Essa Yacoub, and Rainer Goebel. Variable flip angle 3D-GRASE for high resolution fMRI at 7 Tesla. Magnetic Resonance in Medicine, 76(3):897–904, 2016.
[4] Matthias Günther, Koichi Oshio, and David A. Feinberg. Single-shot 3d imaging techniques improve arterial spin labeling perfusion measurements. Magnetic Resonance in Medicine, 54(2):491–498, 2005.
[5] Valentin G Kemper, Federico De Martino, An T Vu, Benedikt A Poser, David A Feinberg, Rainer Goebel, and Essa Yacoub. Sub-millimeter T2 weighted fMRI at 7T: comparison of 3d-GRASE and 2d SE-EPI. Frontiers in neuroscience, 9, 2015
[6] Anagha Deshmane, Vikas Gulani, Mark A. Griswold, and Nicole Seiberlich. Parallel MR imaging. Journal of Magnetic Resonance Imaging, 36(1):55–72, 2012.
[7] John P. Mugler. Improved three-dimensional GRASE imaging with the sort phase-encoding strategy. Journal of Magnetic Resonance Imaging, 9(4):604–612, 1999.
[8] Reed F. Busse, Anja C.S. Brau, Anthony Vu, Charles R. Michelich, Ersin Bayram, Richard Kijowski, Scott B. Reeder, and Howard A. Rowley. Effects of refocusing flip angle modulation and view ordering in 3D Fast Spin Echo. Magnetic Resonance in Medicine, 60(3):640– 649, 2008.
[9] Jorge Jovicich and David G. Norris. GRASE imaging at 3 Tesla with template interactive phase–encoding. Magnetic Resonance in Medicine, 39(6):970–979, 1998.
[10] PJ Beatty, AC Brau, S Chang, SM Joshi, CR Michelich, E Bayram, TE Nelson, RJ Her- fkens, and JH Brittain. A method for autocalibrating 2-D accelerated volumetric parallel imaging with clinically practical reconstruction times. In Proc Intl Soc Magn Res Med, volume 15, page 1749, 2007.
[11] AC Brau, PJ Beatty, S Skare, and R Bammer. Efficient computation of autocalibrating parallel imaging reconstruction. In Proceedings of the 14th Annual Meeting of ISMRM, page 2462, 2006.
Figures