1470
Determining the Time Efficiency of Quantitative MRI Methods using Bloch Simulations1Quantitative Imaging, Delft University of Technology, Delft, Netherlands, 2Biomedical Imaging Group, Erasmus Medical Center, Rotterdam, Netherlands, 3Radiology, Academic Medical Center, Amsterdam
Synopsis
When measuring $$$T_1, T_2, T_2^*, PD$$$, or $$$B_1^+$$$, we prefer the MRI sequence that provides the best precision in the allowed scan time (i.e. having optimal time efficiency). However, experimentally determining the time efficiency is impractical when comparing many sequences, each possibly with varying settings, and multiple tissue types of interest. Here, we derive time efficiency through Bloch simulations which is applicable to any MRI sequence and tissue type. A specific strength of our framework is that it does not require an explicit fitting procedure which may not yet exist when designing novel MR sequences.
Purpose
Presenting and demonstrating a method to determine the time efficiency of quantitative MR sequences. This method is based on Bloch simulations and should be applicable to any MR sequence and tissue type in order to facilitate experimental design and to enhance theoretical insight
Methods
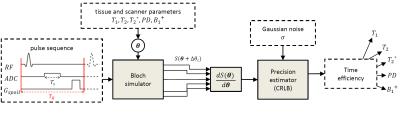
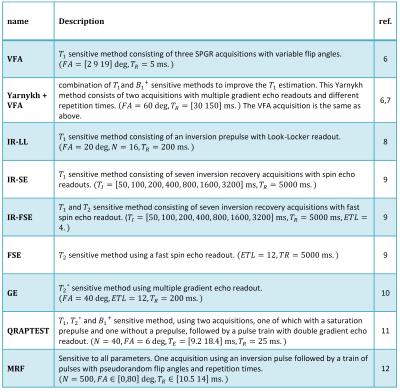
Quantitative MRI (qMRI) methods conventionally measure the proton density, $$$PD$$$, spin-lattice relaxation time, $$$T_1$$$, spin-spin relaxation time, $$$T_2$$$, and the apparent spin-spin relaxation time, $$$T_2^*$$$. This often requires knowledge of the (local) radiofrequency transmit field scaling, $$$B_1^+$$$. For a given qMRI pulse sequence and parameters $$$\theta:=(T_1, T_2, T_2^*, PD, B_1^+)$$$, the Bloch equations describe the expected MRI signal, $$$S(\theta)$$$. The information that $$$S(\theta)$$$ contains on the underlying parameters can be expressed by the Fisher matrix 1: $$\mathcal{I}(\theta) = \frac{1}{\sigma^2} \left(\frac{\partial S(\theta)}{\partial \theta}\right)^T \left(\frac{\partial S(\theta)}{\partial \theta }\right) \in \mathbb{R}^{5\times 5}, $$ where $$$\sigma $$$ equals the noise level. The Cramér-Rao lower bound (CRLB) theorem states that $$$\Sigma(\theta ):=\mathcal{I}(\theta )^{-1}$$$ is a lower bound on the covariance matrix of any unbiased estimator of $$$ \theta$$$ 1. We assume that for each qMRI sequence, we can construct such an unbiased estimator of $$$\theta$$$ that attains the CRLB, for instance by using a maximum likelihood estimator 2,3. Accordingly, we define the precision with which a parameter $$$\theta_i$$$ can be estimated by $$$\Sigma_{i,i}(\theta )^{-1}$$$. The upper limit of the precision is given by $$$\mathcal{I}_{i,i}(\theta)$$$. However, this bound is only reached when the signal response to a change in $$$\theta_i$$$ is not correlated to changes induced by the other MR parameters. The precision is normalized to compensate for differences in tissue parameters and scan time: $$\bar{\mathcal{I}}(\theta):=D^T\mathcal{I}(\theta)D/T_{acq} + P \in \mathbb{R}^{5\times 5},$$ where $$$T_{acq}$$$ equals the scan time, and $$$D$$$ and $$$P$$$ are diagonal matrices that respectively contain reference values of $$$\theta$$$ and a small amount of prior knowledge to ensure $$$\bar{\mathcal{I}}(\theta)$$$ is invertible. We define the time efficiency with which a parameter can be estimated as $$$\theta_i$$$ as $$\mathcal{E}_i(\theta) := \left(\bar{\Sigma}_{i,i}(\theta)\right)^{-1} \in \mathbb{R}.$$ $$$S(\theta)$$$ was obtained with Bloch simulation 4 for a variety of qMRI methods, each modeled as ordered sequences of RF pulses, gradient waveforms, readout time, and delays, see Table 1. Both the RF and gradient pulses were modeled as instantaneous effects. In order to simulate $$$T_2’$$$ decay, the signal was based on an ensemble of spins with a Cauchy distribution of off-resonance frequencies 5. The k-space sampling strategy was not modelled as our time efficiency measure is invariant to the chosen k-space trajectory. Figure 1 shows an overview of our framework. Each sequence was simulated for white matter ($$$T_1/T_2/T_2^* = 617/78/59$$$ ms) and muscle tissue ($$$T_1/T_2/T_2* = 1008/44/30$$$ ms), with $$$PD=1000$$$ and $$$B_1^+=1$$$. For each parameter $$$\theta_i$$$, we set $$$D_{i,i}$$$ to the given value, and $$$P_{i,i}=10^{-8}$$$.Results
Figure
2 shows high time efficiency when qMRI methods estimate the parameters for
which they were originally designed. Exceptions were the VFA and IR-LL methods for $$$T_1$$$ estimation. Improved
time efficiency can be observed by including $$$B_1^+$$$ measurements in VFA. Generally,
the time efficiency of a parameter improved dramatically, when assuming the
other parameters are known (compare $$$\bar{\mathcal{I}}_{i,i}(\theta)$$$, white,
with $$$\mathcal{E}_i(\theta)$$$, colored). Within the presented selection of
methods, maps of multiple parameters are best obtained using QRAPTEST or MRF,
while single parameters may be best determined using IR-FSE $$$(T_1)$$$, FSE
$$$(T_2)$$$, or GE $$$(T_2^*)$$$. Time efficiency values show a small decrease
when moving from white matter to muscle tissue which has a lower expected SNR.Discussion
The results demonstrate that our framework is applicable to a range of qMRI sequences and tissues. Furthermore, the derived time efficiency measure is useful for sequence comparison. Clearly, some methods (e.g. VFA) cannot provide precise estimation without knowledge on the other parameters ($$$B_1^+$$$). A limitation of our work is that by normalizing with the time duration, differences in the minimally scan time required in practice are not considered. Furthermore, we do not model factors such as subject movement and blood flow. Therefore, future research will evaluate if the differences in time efficiency match actual time efficiency differences in the scanner. In this work we show a selection of methods in order to introduce our time efficiency measure. However, the proposed framework can test any qMRI sequences and settings in sillico, without requiring a costly scanner implementation.Conclusion
Our framework for determining the time efficiency of quantitative MRI sequences is applicable over a wide range of sequences, settings, and tissue types. As such, it is a versatile, practical tool for designing qMRI experiments and theoretical insight into MR sequences.Acknowledgements
No acknowledgement found.References
1. Van den Bos A. Parameter estimation for scientists and engineers. John Wiley & Sons; 2007.
2. Karlsen OT, Verhagen R, Bovée WMMJ. Parameter estimation from Rician-distributed data sets using a maximum likelihood estimator: Application to T1 and perfusion measurements. Magn. Reson. Med. 1999;41:614–623
3. Sijbers J, den Dekker A J. Maximum likelihood estimation of signal amplitude and noise variance from MR data. Magn. Reson. Med. 2004;51:586–594.
4. Benoit-Cattin H, Collewet G. Numerical implementation of the Bloch equations to simulate magnetization dynamics and imaging. Proceedings of the 14th Scientific Meeting of the ISMRM; 2006.
5. Stöcker T, Vahedipour K, Pflugfelder D, Shah NJ. High-performance computing MRI simulations. Magnetic Resonance in Medicine 2010;64:186–193.
6. Cheng H-LM, Wright GA. Rapid high-resolution T1 mapping by variable flip angles: Accurate and precise measurements in the presence of radiofrequency field inhomogeneity. Magn. Reson. Med. 2006;55:566–57.
7. Yarnykh VL. Actual flip-angle imaging in the pulsed steady state: A method for rapid three-dimensional mapping of the transmitted radiofrequency field. Magn. Reson. Med. 2007;57:192–200.
8. Kay I, Henkelman RM. Practical implementation and optimization of one-shot T1 imaging. Magn. Reson. Med. 1991;22:414–424.
9. Deoni SCL, Rutt BK, Peters TM. Rapid combined T1 and T2 mapping using gradient recalled acquisition in the steady state. Magn. Reson. Med. 2003;49:515–526.
10. Rossi C, Boss A, Haap M, Martirosian P, Claussen CD, Schick F. Whole-body T2* mapping at 1.5 T. Magnetic Resonance Imaging 2009;27:489–496.
11. Warntjes JBM, Dahlqvist O, Lundberg P. Novel method for rapid, simultaneous T1, T2*, and proton density quantification. Magn Reson Med 2007;57:528–537.
12. Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, Griswold MA. Magnetic resonance fingerprinting. Nature 2013;495:187–192.
Figures