1453
A graphical programming environment for creating and executing adaptive MRI protocols1Diagnostic and Interventional Imaging, University of Texas Health Science Center at Houston, Houston, TX, United States, 2Texas Advanced Computing Center, University of Texas at Austin, Austin, TX, United States, 3Philips Healthcare, Cleveland, OH, United States
Synopsis
Inline processing of magnetic resonance images allows fast feedback of analysis and immediate access to quantitative information. It further allows the development of adaptive MRI protocols. Here, we present GRAPE, a development platform for graphical programming to facilitate the development of adaptive magnetic resonance imaging (MRI) protocols. This platform provides tools to enable graphical creation, execution, and debugging of image analysis algorithms integrated with the MRI scanner, all within a graphical environment. GRAPE is demonstrated with the implementation of patient-specific optimization of the scan parameters of 3D fluid-attenuated inversion recovery (FLAIR) protocol to enhance the contrast of brain lesions in multiple sclerosis, performed on a 3.0 Tesla MRI scanner.
INTRODUCTION
Several medical image analysis and visualization packages, algorithms, and libraries have been developed over the years1–3, and graphical user interface tools have been developed for the design of analysis pipelines.4,5 However, majority of these package and frameworks mainly focused on off-line analysis. Real-time analysis and integration of the analysis pipelines with MRI data acquisition, a critical component of personalized imaging, have not so far been addressed. To fill this gap, we present an extensible GRAphical Pipeline Environment (GRAPE) for the design and execution of analysis pipelines in an adaptive scan environment. GRAPE is designed to provide real-time feedback, enable synchronization of image analysis with data acquisition, provide a user-friendly framework for adaptive protocols, and communicate data with the MRI scanner. GRAPE provides a flexible graphical cross-platform environment that allows users to implement new algorithms, or port existing ones. We describe GRAPE and, as an example, demonstrate its application for patient-specific optimization of fluid-attenuated inversion recovery (FLAIR) imaging for the assessment of brain lesions in multiple sclerosis.METHODS
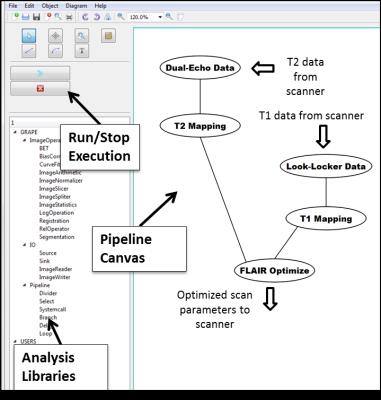
The GRAPE Framework: GRAPE is implemented using the Qt C++ application development framework. GRAPE (Fig. 1) provides an interactive interface that allows the user to create and edit various modules in the analysis pipeline, and to control the data flow. Basic libraries are included in GRAPE for reading, processing of images, and saving output data. Image display modules can display 2D, 3D, or 4D images that allow the user to review intermediate results, particularly useful during initial development. Logical operations, list selection, branching, and looping tools are provided as ready-to-use modules. The image processing library includes various modules for image arithmetic, slicing, statistics, and comparison. GRAPE incorporates widely-used tools such as brain extraction, bias correction, image registration and segmentation. These modules can be used directly by users without any need for programming. External user programs and image analysis functionalities in third party applications can be accessed through command line interface module, providing flexibility and eliminating need for re-programming, allowing quick prototyping of new pipelines. Commonly used image formats such as DICOM, Analyze, and NIFTI are supported. Users can implement new modules by coding the functional components of the module: input assignment, output assignment, validation, and execution. Once a pipeline is created, the user can initiate and stop the execution of the pipeline through the graphical interface or from command line. During execution, color cues indicating the current progress of the pipeline are shown for each module. GRAPE was successfully built under Windows, Linux, and Mac environments, and was run on both standalone computers as well as on the Stampede Linux cluster (Texas Advanced Computing Center, https://www.tacc.utexas.edu/stampede).
Scanner Integration: GRAPE was integrated with a 3.0 T Philips Ingenia MRI scanner (Philips, Best, The Netherlands). A special tool provides interface to the scanner database for automated image extraction immediately after data acquisition. and for data transfer to the GRAPE analysis environment. Input files are handled by a Source module. Encountering an End signal in GRAPE triggers automatic transfer of the pipeline outputs back to the scanner, including image data or scan parameter files. The images are then automatically imported back into the scanner database, and become available for review on the scanner or via transfer to PACS. The whole process of data export, transfer, GRAPE processing, and importing results is fully automated and performed in real time.
RESULTS
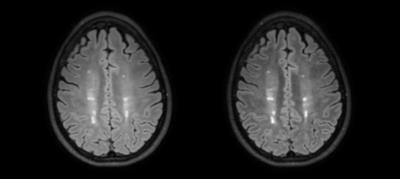
To demonstrate the functionality of GRAPE, a pipeline was developed for patient-specific optimization of FLAIR. The pipeline aims to improve the conspicuity of brain white matter lesions in patients with multiple sclerosis. The inversion time and echo time of FLAIR were optimized for each patient based on fast mapping of the relative proton density (PD) and T1 and T2 relaxation times of the brain tissues (Fig. 1). Programs performing PD/T2 mapping, T1 mapping, and the optimization of the scan parameters of FLAIR were previously developed for scripted execution, and were easily imported in the pipeline using the command line module, avoiding the need for re-programming (Fig. 1). This pipeline was designed such that PD/T2 processing occurred during the acquisition of the T1-mapping pulse sequence, thus reducing the overall time delay by ~46 sec (25%). The optimized scan parameters were subsequently applied to acquire FLAIR in the same scan session, and improved lesion-brain contrast, compared to the fixed-parameter protocol, was observed (Fig. 2).CONCLUSION
GRAPE is a graphical pipeline programming environment that allows rapid development and execution of inline and real-time image analysis. As an example, GRAPE was demonstrated for building a patient-optimized FLAIR protocol.Acknowledgements
This work was supported by the Clinical Translational Science Award (CTSA) Grant UL1 TR000371 from the e National Institutes of Health (NIH) National Center for Advancing Translational Sciences, and the Chair in Biomedical Engineering Endowment Funds. Stampede was generously funded by the National Science Foundation (NSF) through award ACI-1134872. We thank Xiaojun Sun for useful discussion and Vipulkumar Patel for help with the MRI experiments.References
1. Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–173.
2. Friston KJ, Holmes a. P, Worsley KJ, Poline J-P, Frith CD, Frackowiak RSJ. Statistical parametric maps in functional imaging: A general linear approach. Hum Brain Mapp. 1995;2:189–210.
3. Ibanez L, Schroeder W, Ng L, Cates J. The ITK Software Guide [Internet]. http://www.itk.org/ItkSoftwareGuide.pdf; 2005. Available from: http://www.itk.org/ItkSoftwareGuide.pdf
4. Rex DE, Ma JQ, Toga AW. The LONI Pipeline Processing Environment. Neuroimage. 2003;19:1033–1048.
5. Zwart NR, Pipe JG. Graphical programming interface: A development environment for MRI methods. Magn Reson Med. 2014;74:1449–1460.
Figures