1452
Rapid registration of EPI to high-resolution structural images1Department of Biomedical and NeuroMotor Sciences, University of Bologna and Functional MR Unit, Policlinico S. Orsola - Malpighi, University of Bologna, Bologna, Italy
Synopsis
Post-processing methods that can non-linearly register diffusion-weighted EPI data to high-resolution images are useful in the context of clinical protocols. The current research attempts to apply currently available registration methods to rapidly perform such registration. Investigations on healthy subjects show that an appropriately generated target image allows good quality registration to be performed even with freely available software. This is useful for example to provide cortical seeds for diffusion tractography.
INTRODUCTION
Different MRI sequences are susceptible to local variations in static field gradients to different degrees, leading to the problem of spatial mis-registration, for example when comparing high-resolution T1-weighted (T1W), and EPI images. Although corrections can be applied “at the scanner”, by acquiring multiple phase orientations,1 or field maps,2 post-processing registration techniques continue to be useful, but can be computer intensive. The current research attempts to apply currently available registration methods to rapidly register subjects’ diffusion-weighted EPI data to their own T1W volume.METHODS
Twenty-five healthy adult subjects (13 females) underwent MR scanning using a 1.5 Tesla GE Signa system. A T1-weighted axial volumetric image was acquired using the FSPGR sequence (25.6 cm2 FOV; 1 mm isotropic voxels) and axial diffusion-weighted images using a single-shot SE-EPI sequence with FOV 32x32 cm, 3 mm slice-thickness, in-plane resolution 128x128, b-value = 900 s mm-2, 64 diffusion-weighted directions + 7 un-weighted scans. The protocol was approved by the local Ethics Committee and written informed consent was obtained from all participants.
Initial processing for each image included brain extraction (using FSL3 tool BET, and AFNI4 tool 3dSkullStrip), intensity correction (using FSL FAST), while DWI data were processed to correct eddy current and motion effects (FSL FLIRT), and yield DTI parameters, using FSL DTIFIT. Mean of scans without diffusion encoding was calculated (T2W).
Several methods of registration were attempted:
1. No registration. T1W resampled to EPI resolution.
2. Linear registration of T1W image to T2W (FSL FLIRT).
3. M2 + non-linear registration of T1W to T2W using ART5,6 unwarp2d, to restrict warping to EPI scan plane.
4. M2 + non-linear registration using AFNI 3dQwarp, following recommended protocol.
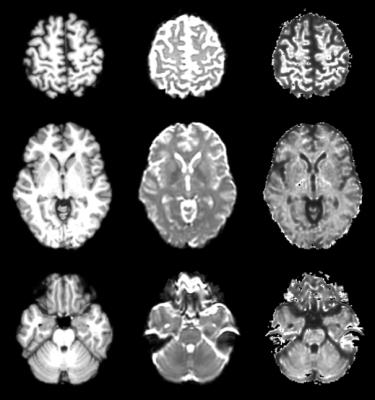
5. As M2, but using a synthetic image based on DTI eigenvalues, and exhibiting more T1-like contrast, in place of T2W (Figure 1).
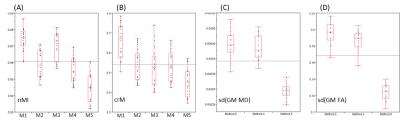
To assess the performance of each method, aside from visual inspection, global similarity measurements (normalised mutual information and symmetrised correlation ratio) were calculated for each image pair using AFNI tool 3dAllineate. In addition, variability of MD and FA values were calculated for grey matter voxels, after registration methods 2, 4, and 5.
RESULTS
Image warping using 3dQwarp took 1’ 40” on average, while applying the warp to the DWI volume took another 5’ 09”. Using unwarp2d, warping time was 5 s, and 4’ 17” was needed to apply the warp to the DWI data.
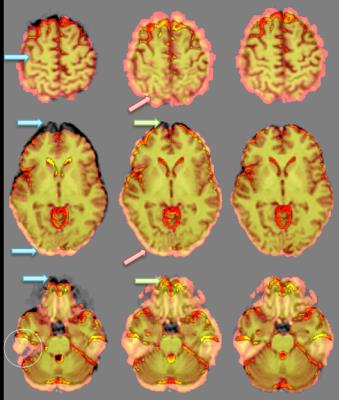
Visual assessment showed that the matching of brain features on high-resolution and EPI images was globally improved after simple affine registration, but due to the global warping induced in the EPI image due to field shifts, improvement in some areas was balanced by deterioration in others (Figure 2). In-plane non-linear warping using unwarp2d, using a synthetic image as a target, resulted in visibly better feature matching (Figure 2). Using the T2W EPI image resulted in a worse image than linear registration alone.
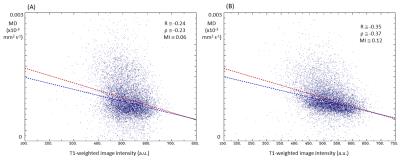
These findings are reflected in the global similarity metrics (Figure 3). Note especially that variance in both MD and FA are reduced after Method 5, reflecting the fact that that selected tissue did not extent so far into either regions of CSF or white matter, compared to either Method 2 or Method 4. The improved match is also confirmed by an inspection of the scatter plot of MD against T1W image intensity (Figure 4).
DISCUSSION
Of the most commonly used MR non-linear registration tools, FNIRT (FSL) and DARTEL (SPM7) are not designed for registration between images with greatly differing contrast. The solution provided by AFNI (3dQwarp) does improve on affine registration, although it is somewhat slow. The starting point provided by FLIRT may be sub-optimal, and this may have affected the performance of the algorithm. While the ART package is less well known, its performance in the context of intra-modal registration has been noted by previous researchers.8 An attempt to directly warp participants’ T2W EPI images to their T1W images did not yield good results (Method 3). However, using the rotation-invariant information available in the processed DWI data to create a synthetic image with contrast more similar to the T1W data did allow the ART unwarp2d algorithm to perform at least as well as 3dQwarp. Because most distortion in EPI images occurs in the imaging plane, the algorithm is particularly suited to this kind of registration.CONCLUSION
Rapid registration of EPI to high-resolution MR images is possible with freely available software tools. The synthetic image used has not yet been optimised and future work will attempt to improve the match between the image generated and the high-resolution target.Acknowledgements
No acknowledgement found.References
1. Andersson JLR, Skare S, Ashburner J. NeuroImage, 20(2):870-888, 2003.
2. Jezzard P, Balaban RS. 1995. MRM. 1995 Jul;34(1):65-73.
3. FMRIB Software Library v5.0. http://fsl.fmrib.ox.ac.uk/fsl/fslwiki. Accessed October 9 2016.
4. National Institutes of Health – AFNI. https://afni.nimh.nih.gov. Accessed October 9 2016.
5. Ardekani BA, Braun M, Hutton BF, Kanno I, Iida H. A fully automatic multimodality image registration algorithm. J Comput Assist Tomogr. 1995 Jul-Aug;19(4):615-23.
6. Automatic registration toolbox. http://www.nitrc.org/projects/art. Accessed October 9 2016.
7. Klein R, Andersson J, Ardekani BA, et al. Neuroimage. 2009 July 1; 46(3): 786–802
8. SPM. http://www.fil.ion.ucl.ac.uk/spm/software/spm8. Accessed October 9 2016.
Figures