1451
Quantitative and qualitative evaluation of bias field correction methods1Biomedical Magnetic Resonance, Otto-von-Guericke University, Magdeburg, Germany, 2Center for Behavioral Brain Sciences, Magdeburg, Germany, 3Leibniz Institute for Neurobiology, Magdeburg, Germany, 4German Center for Neurodegenerative Disease (DZNE), Magdeburg, Germany
Synopsis
Bias field correction is an essential prerequisite for image analysis, especially at high field strength. In this study multiple correction methods, based on acquisition of a reference image and computational approaches with varied input parameters, are compared. The results indicate that acquisition of a conventional MPRAGE corrected by SPM yields quantitatively and qualitatively comparable results to acquiring a reference image (e.g. MP2RAGE), however, scan time is up to halfed.
Introduction
Bias field correction is a crucial pre-processing step as it affects qualitative and quantitative image analysis, especially at high field strength. Computational techniques can estimate the bias field from the image itself, e.g. the widely used “nonparametric nonuniform intensity normalization” (N3)1, its evolution the N4 algorithm2, and SPM’s bias field correction method “unified segmentation”3. Alternatively, acquiring a reference image enables correction by division4 (referred to DIV in this work), which was incorporated into the “MP2RAGE” sequence5. MP2RAGE facilities sophisticated combination methods of the two simultaneously acquired images compared to the simple correction by division. However, both methods approximately double the scan time compared to a conventional MPRAGE6 with the same parameters. Therefore, we compare in this study computational techniques with a large variety of parameters against methods based on the acquisition of reference images to remediate the bias field.
Methods
One healthy, highly experienced male Caucasian subject (born 1982) participated in this study, which was approved by the local ethics committee. The subject gave written informed consent prior to participation.
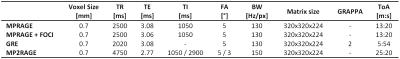
Scanning has been conducted on a whole body 7T MR scanner (Siemens Healthcare, Erlangen, Germany) using a 32-channel receive head coil (Nova Medical Head Coil, Siemens, Erlangen, Germany). Image data was acquired using the MPRAGE sequence, with the standard and TR-FOCI inversion pulse7, the MP2RAGE sequence and GRE sequence. The isotropic resolution of all datasets is 700µm and TI was identical in all protocols (Tab.1). Except for the MP2RAGE all volumes were acquired using prospective motion correction8,9. The subject was fixated in the head coil using foamed cushions and the MP2RAGE was acquired first to reduce inevitable motion during the measurement as much as possible. Parameters of N3, N4 and SPM’s bias field correction were systematically varied. In order to assess the quality of bias field correction the coefficient of variation (CV) of the intensity of white matter (WM), grey matter (GM) and a joint variation of between both was determined.
$$CV_T=σ_T/µ_T $$
$$CV_J=(σ_{WM}+σ_{GM})/|μ_{WM}-μ_{GM}|$$
Where T stands either for WM or GM and σ and µ denote the standard deviation and mean intensity of the tissue, respectively. To extract tissue classes all datasets were masked using the same white and grey matter segmentation created by FreeSurfer10 after inhomogeneity correction.
Results
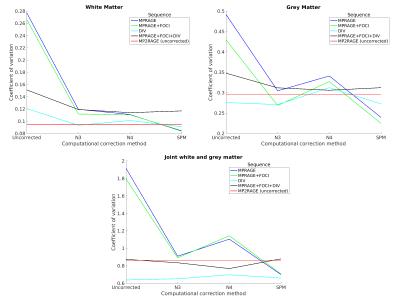
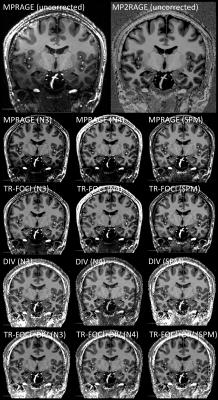
After systematic variation of the parameters of the N3, N4 and SPM’s algorithm (not shown), the best combination for 7T data was identified (Tab.2). The CV was estimated for all datasets and compared to determine the best correction method quantitatively (Fig.1) and qualitatively (Fig.2).Discussion
Qualitatively the bias field correction is very similar. However, a bit more signal in the temporal lobes could be retrieved with the correction methods based on a reference image (Fig.2). Though, the signal of the temporal lobes is low in all datasets, making it probably difficult for automatic segmentation methods to identify tissue classes correctly.
The coefficient of variation was used to assess the quality of the bias field correction quantitatively. A potential drawback is that it is prone to image noise11. Therefore, images were acquired at 7T without any acceleration or partial Fourier acquisition and with low bandwidth to maximize signal-to-noise ratio. The results suggest overall good homogeneity of WM and GM for all corrected datasets. The coefficient of joint variation is lowest in case of the correction methods using DIV and further correcting it with SPM. Hence, the intensity variability within the tissue classes decreases, while maintaining a good separation between mean tissue intensities and suggests a benefit in further image analysis, e.g. segmentation methods. If corrected with SPM the difference in joint variation of the DIV compared to the conventional MPRAGE or MPRAGE with TR-FOCI amounts to 6% and 7%, respectively. Visually the MPRAGE and MPRAGE with TR-FOCI are very similar with a slight advantage of the TR-FOCI in the temporal lobes.
The CV of WM and GM in case of acquisition of an MP2RAGE can potentially be improved if the computational correction methods are also applied to it. A maximum degradation of 10% in joint variation was determined using the correction by division and further computational correction though. The parameters of the N4 algorithm probably needs to be further adjusted to yield at least the same quantitative results as its predecessor N3.
To conclude, the results suggest that the bias field correction with SPM using a conventional MPRAGE (with TR-FOCI) yields best results, if quantitative T1-values are not specifically needed, because it can be acquired faster than using a correction method based on a reference image, while having qualitatively and quantitatively very similar results.
Acknowledgements
This work was supported by the NIH, grant number 1R01-DA021146, and by the Initial Training Network, HiMR, funded by the FP7 Marie Curie Actions of the European Commission, grant number FP7-PEOPLE-2012-ITN-316716.References
1. Sled, J. G., Zijdenbos, A. P. & Evans, A. C. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE transactions on medical imaging 17, 87–97 (1998).
2. Tustison, N. J. et al. N4ITK: improved N3 bias correction. IEEE transactions on medical imaging 29, 1310–1320 (2010).
3. Ashburner, J. & Friston, K. J. Unified segmentation. NeuroImage 26, 839–851 (2005).
4. van de Moortele, P.-F. et al. T1 weighted brain images at 7 Tesla unbiased for Proton Density, T2* contrast and RF coil receive B1 sensitivity with simultaneous vessel visualization. Neuroimage 46, 432–446 (2009).
5. Marques, J. P. et al. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. Neuroimage 49, 1271–1281 (2010).
6. Mugler, J. P. & Brookeman, J. R. Three-dimensional magnetization-prepared rapid gradient-echo imaging (3D MP RAGE). Magn. Reson. Med. 15, 152–157 (1990).
7. Hurley, A. C. et al. Tailored RF pulse for magnetization inversion at ultrahigh field. Magnetic resonance in medicine 63, 51–58 (2010).
8. Stucht, D. et al. Highest Resolution In Vivo Human Brain MRI Using Prospective Motion Correction. PloS one 10, e0133921 (2015).
9. Maclaren, J. et al. Measurement and correction of microscopic head motion during magnetic resonance imaging of the brain. PloS one 7, e48088 (2012).
10. Fischl, B. FreeSurfer. NeuroImage 62, 774–781 (2012).
11. Chua, Z. Y., Zheng, W., Chee, M. W. L. & Zagorodnov, V. Evaluation of performance metrics for bias field correction in MR brain images. Journal of magnetic resonance imaging : JMRI 29, 1271–1279 (2009).
Figures