1443
Optimization of 2D registration using minctracc on myelin stained brain slices1Department of Radiologic Technology, Carinthia University of Applied Sciences, Klagenfurt, Austria, 2Institute for Applied Research on Ageing, Carinthia University of Applied Sciences, Klagenfurt, Austria, 3Center for Brain Research, Medical University of Vienna, Vienna, Austria, 4Medical Physics Group, Institute for Diagnostic and Interventional Radiology, Jena University Hospital – Friedrich Schiller-University, Jena, Germany, 5Centre for Advanced Imaging, University of Queensland, Brisbane, Australia, 6Montreal Neurological Institute, McGill University, Montreal, Canada, 7University Clinic for Trauma Surgery, Medical University Vienna, Vienna, Austria, 8High Field Magnetic Resonance Centre, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 9Department of Medical Engineering, Carinthia University of Applied Sciences, Klagenfurt, Austria, 10Center of Anatomy and Cell Biology, Medical University Vienna, Vienna, Austria
Synopsis
Histological analyses are important for a wide spectrum of in vivo and in vitro imaging projects. But unlike MRI or CT, histological analyses are typically performed in two dimensions. Nonlinear tissue deformation and ruptures of brain tissue are often common, making analysis in slice direction more difficult. In this work, we optimized a hierarchical, nonlinear fitting pipeline on the basis of two high resolution, myelin stained brain sections using mintracc.
Purpose:
Modern 3D, in vivo and in vitro imaging of the human brain is of high value in research and clinical applications. In many cases, a combination with histological analysis is important. As histological analyses are performed in two dimensions, a comparison between neighboring sections can be very challenging as tissue preparation and analysis (fixation, dehydration, cut in the microtome, staining ..) can cause nonlinear tissue deformation and ruptures. The purpose of this work was to optimize image registration on myelin stained brain sections to reduce miss alignment by using minctracc [1], which is part of the MINC- toolbox [2] in order to facilitate 3D analyses of histological data and comparison to 3D MRI data.Methods:
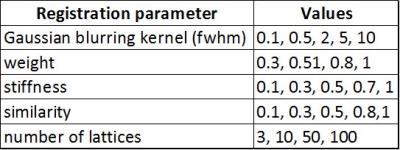
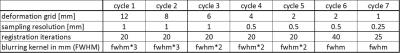
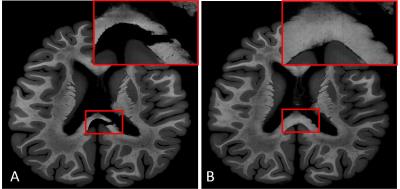
High resolution images (0.03 x 0.03 mm) of two consecutive myelin stained brain sections with a thickness of 10µm were selected as source and target for nonlinear registration optimization. These two images (Figure 1) were chosen as they show a moderate amount of nonlinear distortion and a clearly visible rupture in the corpus callosum. Both images were manually masked to eliminate background noise, intensity corrected [3] and linear aligned using mri_robust_register [4] prior to nonlinear registration. To find the optimal registration parameters for this contrast and resolution, all combinations (2000) of the registration (minctracc) parameters shown in Table 1 were applied to the hierarchical, nonlinear fitting pipeline described in Table 2. Mintracc was used with the magnitude of the blurred input data and correlation coefficients were used as objective function. Within the hierarchical fitting process, fitting parameters are successively changed to avoid local minima and to increase fitting accuracy. For registration evaluation, two ROIs were defined in the target image, one in the area of the corpus callosum tear and one in the remaining brain tissue. The registered source image and the target image were binarized (intensities > 0 were set to 1) and logically compared using an “exclusive or” function as similarity measure. The sum of pixels representing logical ones (LO) was used to evaluate registration accuracy. Additionally, we analyzed the best 200 registration results (10%) in order to show a parameter tendency towards accurate registration.Results:
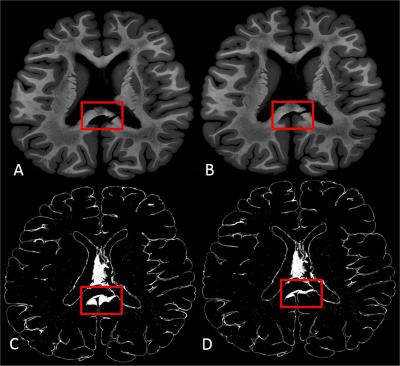
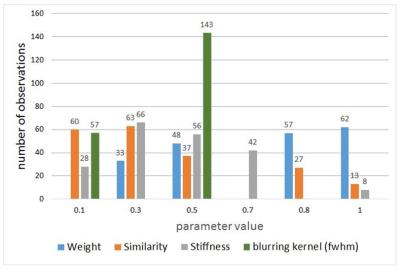
Registration parameter optimization is crucial in order to obtain good registration results. The range of LOs for different registration parameters ranged between 26870 and 185849 for the ROI around the corpus callosum rupture and between 202049 and 4280029 for the remaining tissue. Figure 2 shows two registration results for different registration parameters. Over all, we found the following parameters to be best suited for the actual data: stiffness=0.3, similarity=0.1; number of lattices=100; weight=1, blurring kernel (fwhm)=0.5mm. Figure 3 displays a histogram of the registration parameters for the best 200 registration results. As can be seen, the weighting shows a trend to higher numbers (0.8 – 1), the similarity and stiffness tend to numbers around 0.3 and the filter kernel with a fwhm of 0.5 mm was found most often in the 200 best registration results. Note that the filter kernel is changed during the hierarchical fitting process. In the 200 best registration results, 71 times a lattice of 10, 66 times a lattice of 100 and 63 times a lattice of 50 was found.Discussion:
As demonstrated in this work, optimization of registration parameters is important in order to increase registration accuracy, especially for 2D data. The determined best fitting parameters showed a trend towards specific values, but unfortunately it is not possible to present the “perfect” fitting parameters as they are highly dependent on the input data. It is also important to be aware that too large deformations can cause unrealistic tissue deformations e.g., using other registration parameters the rupture in Figure 1(A) can nearly be closed, however at the expense of large and unrealistic tissue deformation.Acknowledgements
This work was supported by the Österreichische Nationalbank Anniversary Fund project no. 16153References
1) Collins D.L., et al. (1994) Automatic 3D Inter-Subject Registration of MR Volumetric Data in Standardized Talairach Space, Journal of Computer Assisted Tomography, 18(2) pp. 192-205
2) Vincent R.D., et al. (2004) A modality independent format for multidimensional medical images. Proceedings of the 10th Annual Meeting of the Organization for Human Brain Mapping
3) Sled J.G., et al. (1998) A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 17:87–97. doi: 10.1109/42.668698
4) Reuter M., et al. (2010) Highly Accurate Inverse Consistent Registration: A Robust Approach. NeuroImage 53(4), pp. 1181-1196, 2010.
Figures