1437
Optimal contrast enhancement of blockface images for MRI guided reconstruction of mouse brain volumes1Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 2Centre for Advanced Imaging, University of Queensland, Brisbane, Australia
Synopsis
Blockface imaging can improve atlases of the rodent brain by supplying high resolution images. This study compares three different contrast stretching methods for enhancing the information in blockface images together with a registration to a 16.4 T atlas of the mouse brain. Contrast enhancement technique used was histogram equalization, adjusting the image intensity values by stretching them between the bottom 1% and the top 1% and CLAHE with a clip limit of 0.01 and a uniform histogram. Registrations was rigid, affine and SyN. By using CLAHE as contrast stretching method a high similarity to the MRI was found.
Purpose
Blockface imaging is a technique that is usually used as an intermediate when registering histological sectioning to MRI 1. Despite blockface imaging typically not matching the contrast in histological data, it still plays an important role in supplying high-resolution images that can improve atlases of the brain 2. Blockface image stacking has been performed within several studies 3,4 but the exploitation of the greater contrast which can be found within the images together with a registration to MRI volumes has, to the authors knowledge, not been performed. This study investigates three different contrast enhancement techniques together with a registration to a 16.4 T atlas of the mouse brain. The aim of this study was to find the technique that resulted in the greatest contrast for matching the two modalities of blockface imaging and MRI. Code and parameters available here: https://github.com/NIF-au/CERAMethods
Blockface data was obtained from one mouse of the species c57bl/6j wildtype. The images were taken with a NIKON D800E camera and were provided as NEF (Nikon's version of RAW) files. The specimen in the data-set was cut in a transverse plane and the resolution was 4912x7360 with 122 images. MRI data was acquired as an average atlas from Australian Mouse Brain Mapping Consortium (www.imaging.org.au/AMBMC/Model). The MRI data had a resolution of 679x1311x404 with an isotropic voxel size of 0.015mm. Blockface images were converted from RAW to PNG and resized 25%. A semi manual cropping was applied in order to remove the image background. The Otsu's method was subsequently used for global segmentation of the brain, and an automatic linear translation was performed in order to correct the small misalignments in the data set. Contrast stretching was applied to the blockface images by means of stretching the intensity values between the bottom 1% and the top 1% of the pixel values, histogram equalization, and Contrast Limited Adaptive Histogram Equalization (CLAHE) with a clip-limit of 0.01 and a uniform histogram. Finally a sharpening of the image was applied on the selected contrast stretching method. The blockface images were stacked and resampled to create a volume with the resolution of 600x1200x300 and a voxel size of 0.018x0.02x0.02. A resampling of the MRI data was also performed to create the same resolution. Two linear registrations (rigid and affine) were applied to align the outer boundaries of the mouse brain in the blockface images to the MRI. The inner structures of the brain were aligned using Symmetric Normalization (SyN), which is a non-linear diffeomorphic transformation.Results
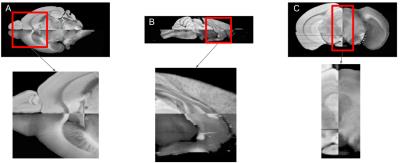
Figure 1 illustrates the different contrast stretching techniques: (A) histogram equalization, (B) adjusting the image intensity values by stretching them between the bottom 1% and the top 1%, (C) CLAHE with a clip limit of 0.01 and a uniform histogram. By visual inspection the greatest contrast enhancement technique was CLAHE. The technique CLAHE with applied sharpening was used for the registration. The results of the final registration between the blockface image and the MRI are illustrated on Figure 2. Figure 2 shows the registration SyN after a rigid and affine transformation were performed ((A) axial, (B) sagittal and (C) coronal views).
Discussion and conclusion
A novel technique for optimal contrast enhancement of blockface images was developed. By visual inspection the technique of CLAHE was chosen due to the highest similarity when comparing to the MRI. The gel and dye used for for embedding the mouse brain in the blockface imaging process resulted in a segmentation that was not completely accurate, indicating that an improvement of the segmentation would be recommended. Blockface imaging with contrast enhancement circumvent the time consuming process of preparing histological sections and has the potential of providing information of the white matter density and the morphology of the brain.Acknowledgements
The authors acknowledge the facilities, and the scientific andtechnical assistance of the National Imaging Facility at the Centrefor Advanced Imaging, the University of Queensland. We would like to thank Otto Møndsteds Fond, Knud Højgaards Fond and Oticon Fonden for travel funds.References
1. Ceritoglu, C., Wang, L., Selemon, L.D., Csernansky, J.G., Miller, M.I. and Ratnanather, J.T., 2010. Large deformation diffeomorphic metric mapping registration of reconstructed 3D histological section images and in vivo MR images. Frontiers in human neuroscience, 4, p.43.
2. Toga, A.W. and Mazziotta, J.C., 2002. Brain mapping: the methods. Academic press
3. Lebenberg, J., Hérard, A.S., Dubois, A., Dauguet, J., Frouin, V., Dhenain, M., Hantraye, P. and Delzescaux, T., 2010. Validation of MRI-based 3D digital atlas registration with histological and autoradiographic volumes: an anatomofunctional transgenic mouse brain imaging study. Neuroimage, 51(3), pp.1037-1046.
4. Alegro, M., Amaro-Jr, E., Loring, B., Heinsen, H., Alho, E., Zollei, L., Ushizima, D. and Grinberg, L.T., 2016. Multimodal Whole Brain Registration: MRI and High Resolution Histology. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops (pp. 194-202).
Figures