1408
Novel annihilation filter framework for accelerated parameter mapping1Electrical and Computer Engineering, University of Iowa, Iowa City, IA, United States
Synopsis
Quantitative parameter maps offer valuable information about various tissue attributes, which are early markers for many neurological disorders. However the long acquisition time of the associated image time series puts a restriction on the achievable spatial resolution. In this work, we introduce a novel framework, which exploits the exponential nature of the time profiles at every pixel and spatial smoothness of the exponential parameters to recover the images from highly under-sampled measurements. Our preliminary results clearly demonstrate the potential of the proposed algorithm.
Purpose
The estimation of exponential parameters (e.g frequency, relaxation parameters) from time series data is a key problem in several MRI applications including parameter mapping, spectroscopic imaging, field mapping, and fat water imaging. The acquisition of multiple images at different settings (e.g. TE, TR, spin-lock duration) is associated with increased scan time, which often restricts the achievable spatial resolution. A common approach to overcome these problems is to acquire under sampled data and regularize the reconstruction using appropriate priors (e.g sparsity low rank etc.).1, 2 However, most of the current regularization priors do not exploit the exponential behavior of the time-series and the spatial smoothness of the parameters. The main focus of this work is to introduce a novel annihilation filter formulation, which directly exploits the exponential structure of the time series and the spatial smoothness of the exponential parameters.Methods
Our work is centered on the linear predictability of a 1-D exponential time-series. This implies that a linear combination of damped/undamped exponentials can be linearly predicted/annihilated by the convolution with a 1-D filter, whose degree is equal to the number of exponentials at that pixel3. We model the temporal signal at each voxel location $$$\mathbf{r}$$$ as $$\rho[\mathbf{r},n] = \sum_{i=1}^{L}\alpha_{i}(\mathbf{r})~\beta_{i}(\mathbf{r})^{n}.$$ In $$$T_2$$$ mapping applications, the exponential parameters $$$\beta_i = \exp\left(\frac{-\Delta T}{T_{2,i}(\mathbf r)}\right)$$$, where $$$\Delta T$$$ is the time between two image frames and $$$T_{2,i}$$$ is the relaxation parameter of the $$$i^{\rm th}$$$ tissue component. Such a signal can be linearly predicted/annihilated by the convolution with a 1-D filter $$$h[\mathbf{r},z]=\prod_{i=1}^{L}(1-\beta_{i}(\mathbf{r})z^{-1})$$$. The exponential parameters often vary smoothly in space; we exploit this fact by constraining the filter coefficients to be spatially band-limited. This results in the following 3-D convolution relation: $$\hat\rho[\mathbf{k},n] \otimes d[\mathbf{k},n] = 0,$$ where $$$\otimes$$$ is the 3-D convolution operator, $$$\rho[\mathbf{k},n]$$$ and $$$d[\mathbf{k},n]$$$ are the k-t space coefficients of the image time-series $$$\rho[\mathbf{r},n]$$$ and the annihilation filter $$$h[\mathbf{r},n]$$$ respectively. The extent of the filter $$$d[\mathbf{k},n]$$$ along the spatial frequency and temporal dimension controls the spatial smoothness of the filters and the exponential parameters respectively. The above annihilation relations can be compactly represented in a matrix form as $$$\mathcal{T}(\hat{\rho})~\mathbf {d} = 0$$$, where $$$\mathcal{T}(\hat{\rho})$$$ is a block Toeplitz matrix and $$$\mathbf d$$$ is the vectorized filter coefficients. See figure (1) for the construction of the Toeplitz matrix. The annihilation relation implies that the matrix $$$\mathcal{T}(\hat{\rho})$$$ is low-rank. Hence, we formulate the recovery of $$$\rho$$$ from undersampled Fourier measurements as the following structured low rank matrix recovery problem in the Fourier domain: $${\mathbf{\hat{\rho}}}^\star = \arg\min_{{\mathbf{\hat{\rho}}}} \|\mathcal{T}(\mathbf{\hat{\rho}})\|_p + \frac{\lambda}{2} \|\mathcal{A}(\mathbf{{\hat{\rho}}}) - \mathbf{b}\|^2_2$$ where $$$\hat{\rho}$$$ are the 2-D Fourier coefficients of the volume $$$\rho$$$, $$$\mathbf{b}$$$ represents the undersampled measurements, $$$\mathcal{A}$$$ is a linear operator that encodes the coil sensitivity and the Fourier undersampling matrices and $$$\lambda$$$ is a regularization parameter. To solve the above equation, we employ an Iterative Reweighted Least Squares (IRLS) based algorithm4.
We demonstrate the algorithm on the recovery of both single channel (coil compressed) and multi-channel data from under-sampled Fourier measurements. For this purpose, a fully sampled 2-D dataset was acquired using a turbo spin echo sequence and the following scan parameters were used: Matrix size - 128x128, Coils = 12, FOV: 22x22 cm2, TR = 2500 ms and slice thickness = 5mm. The $$$T_2$$$ weighted images were obtained for 12 equispaced echo times ranging from 10 to 120ms. Post image recovery, the $$$T_2$$$ maps were estimated by fitting a mono-exponential model to each voxel.
Results
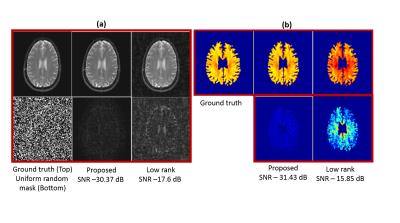
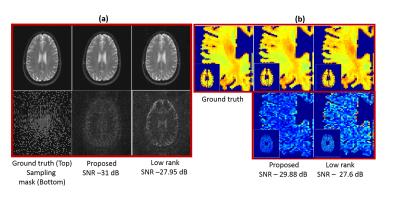
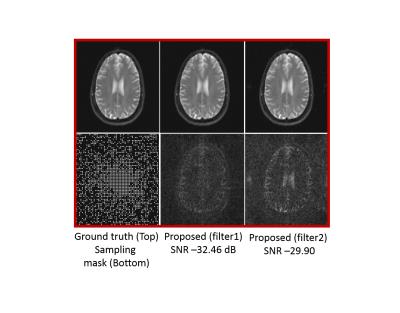
Figure (2) compares the proposed approach with the k-t Low Rank algorithm2 (k-t SLR without the sparsity) on the recovery of single channel data from 30$$$\%$$$ uniform random measurements. In figure (3), we compare the two methods on the recovery of multi-channel data from 12-fold (3-fold variable density + 4-fold 22 Cartesian) undersampled measurements. In both cases, We observe that the reconstructions from the proposed scheme have lower errors (see caption for details). In figure (4), we study the impact of the filter sizes on the quality of the reconstructions. We observe that the errors in the reconstructions are reduced when the spatial support of the filter is decreased, which clearly demonstrates the benefit of exploiting spatial smoothness of the exponential parameters.
Conclusion
We introduced a novel annihilation filter framework to recover the $$$T_2$$$ weighted MR images from under-sampled Fourier measurements. The reconstructions and the $$$T_2$$$ maps from the proposed approach have fewer errors compared to the low rank method. Also, the benefit of exploiting the spatial smoothness of the exponential parameters was demonstrated through the use of smaller filters, which helped improve the reconstruction quality.Acknowledgements
No acknowledgement found.References
1. Lustig, Michael, David Donoho, and John M. Pauly. "Sparse MRI: The application of compressed sensing for rapid MR imaging." Magnetic resonance in medicine 58.6 (2007): 1182-1195.
2. Lingala, Sajan Goud, et al. "Accelerated dynamic MRI exploiting sparsity and low-rank structure: kt SLR." IEEE transactions on medical imaging 30.5 (2011): 1042-1054.
3. Stoica, Petre, and Randolph L. Moses. Introduction to spectral analysis. Vol. 1. Upper Saddle River: Prentice hall, 1997.
4. Mohan, Karthik, and Maryam Fazel. "Iterative reweighted least squares for matrix rank minimization." Communication, Control, and Computing (Allerton), 2010 48th Annual Allerton Conference on. IEEE, 2010.
Figures