1406
Noise and Artifact Reduction in 3D Abdominal and Thoracic Imaging using CAIPIRINHA1Radiology, University of Wisconsin, Madison, WI, United States, 2Medical Physics, University of Wisconsin, Madison, WI, United States, 3Biomedical Engineering, University of Wisconsin, Madison, WI, United States, 4Medicine, University of Wisconsin, Madison, United States, 5Emergency Medicine, University of Wisconsin, Madison, WI, United States
Synopsis
This study investigated Controlled Aliasing In Parallel Imaging Results IN Higher Acceleration (CAIPIRINHA) in combination with Rotated Slab Excitation (ROSE) to reduce parallel imaging artifacts and residual aliasing artifacts using a coronal reconstruction and sagittal excitation. Images acquired using the CAIPIRINHA sampling pattern had fewer parallel imaging artifacts than using a conventional sampling pattern. G-factor analysis also demonstrated improved SNR performance using the CAIPIRINHA sampling pattern. The CAIPIRIHNA sampling pattern when combined with ROSE reduced residual aliasing artifacts and parallel imaging artifacts. This enables higher acceleration factors for shorter scan times, without sacrificing image quality.
Introduction
Coronal 3D T1 weighted imaging is often used with phase encoding in the left/right direction for abdominal imaging applications (e.g. hepatic or renal imaging) and thoracic imaging applications (e.g. magnetic resonance angiography MRA). However, this can cause anatomy (typically the arms) to alias into the field of view. In previous work, Rotated Slab Excitation (ROSE) has been shown to reduce residual aliasing artifacts with coronal abdominal images1. Recently Controlled Aliasing In Parallel Imaging Results IN Higher Acceleration (CAIPIRINHA)2 was shown to be a promising option for maintaining good image quality while reducing scan time with hepatic imaging using a GRAPPA-VIBE sequence3. The purpose of this study was to investigate using CAIPIRINHA in a sequence with ROSE to reduce both parallel imaging artifacts and residual aliasing artifacts with a spoiled gradient echo sequence and sagittal excitation, reconstructed in the coronal orientation.Methods
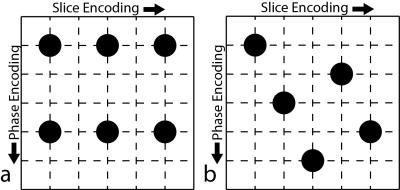
In this study, undersampled 3D-spoiled gradient echo (SGRE) acquisitions with ROSE excitation strategy were performed using conventional undersampling schemes (Figure 1a) and CAIPIRINHA (Figure 1b). Image reconstructions were performed using the Autocalibrating Reconstruction for Cartesian imaging (ARC) reconstruction4. G-factor maps were estimated using the pseudo-multiple replica method5. The performance of the two sequences was then compared in terms of the presence of parallel imaging artifacts, as well as g-factor maps to assess SNR performance.
MRI Measurements
Phantom and in vivo measurements were made on a 3T clinical MRI system (Discovery MR750, GE Healthcare, Waukesha, WI) using a 32-channel torso coil (NeoCoil, Pewaukee, WI). In vivo imaging was performed with IRB approval and informed written consent. A water phantom doped with NiCl2 and NaCl doped-water phantom was scanned with the following parameters: FOV = 36 x 28.8 x 32 cm (X x Y x Z), TE = 1.8 ms, TR = 4.1 ms, 83.33 kHz RBW, 2 mm slice thickness, acquisition matrix of 280x200x200, and the number of autocalibration points was 34x34. Three acquisitions were performed: a reference acquisition with no parallel imaging acceleration and undersampled acquisitions with a conventional (Figure 1a) or CAIPIRINHA (Figure 1b) sampling pattern, both with net acceleration factors of 4.5. The fully sampled reference acquisition took 134.5 seconds while both undersampled acquisitions took 30s. In addition, a noise only scan with bandwidth and receiver gain that are identical to the acquired images was obtained to measure the noise covariance matrix required for estimating the g-factor5.
An example clinical scan was performed for a patient undergoing a chest MRA with contrast. Two post-contrast scans were performed with a coronal acquisition, one with the conventional sampling pattern and the other with the CAIPIRHNA sampling pattern, both with the following parameters: FOV = 44x39.6x33.3 cm, TE = 1.04 ms, TR = 3.1 ms, 83.33 kHz RBW, 1.6 mm slice thickness, acquisition matrix of 280x200x208, and the number of autocalibration points was 34x34.
Results
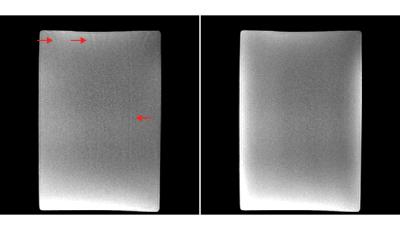
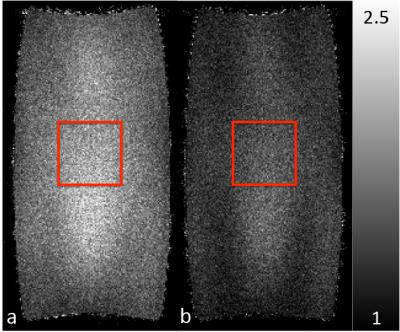
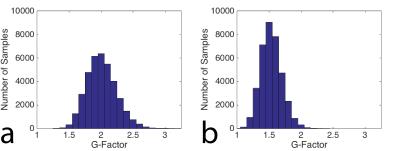
Figure 2 is a coronal slice from the center of the imaging domain acquired using the conventional sampling pattern (Figure 2a) and the CAIPIRIHNA sampling pattern (Figure 2b). The red arrows highlight significant parallel imaging artifacts with the conventional sampling pattern. Figure 3 contains g-factor maps for the sampling patterns. The mean g-factor in a 5x5x5 cm cube in the center of the phantom (highlighted in red in Figure 3) was 2.01 for the conventional sampling pattern and 1.53 for CAIPIRIHNA. Histograms of the g-factor are shown in Figure 4 for the same 3D region of interest (ROI). The histogram for CAIPIRIHNA is clustered at lower g-factors than the conventional sampling pattern, which demonstrates an increase in SNR throughout the entire ROI. Figure 5 contains comparisons of axial reformats of the in vivo scan, showing an artifact that was moved away from the top of the liver and center of the imaging domain with the CAIPIRIHNA sampling pattern.Discussion & Conclusions
The CAIPIRIHNA sampling pattern results in controlled aliasing that moves artifacts to the corners of the domain. This results in fewer parallel imaging artifacts in the center of the imaging domain as well as a smaller parallel-imaging related reduction in SNR compared to the conventional sampling pattern. The CAIPIRIHNA sampling pattern in conjunction with ROSE should reduce residual aliasing artifacts and parallel imaging artifacts and should result in shorter scan times without sacrificing image quality for applications including hepatic imaging or MRA.Acknowledgements
The authors wish to acknowledge support from GE Healthcare, and the National Cancer Institute of the National Institutes of Health under Award Number T32CA009206.References
1. Brau A, Bayram E, Saranathan M, Kawashima A, Rotated Slab Excitation (ROSE) for Reduced Foldover Artifacts in Coronal 3D Abdominal Imaging, In Proceedings of the 16th Annual Meeting of ISMRM, Toronto, Canada, 2008. Page 502.
2. Breuer F, Blaimer M, Mueller M, Seiberlich N, Heidemann R, Griswold M, Jakob P, Controlled Aliasing in Volumetric Parallel Imaging (2D CAIPIRINHA), Magn Reson Med 2006; 55:549-556.
3. Morani A, Vicens R, Wei W, Gupta S, Vikram R, Balachandran A, Reed B, Ma J, Qayyum A, Szklaruk J, CAIPIRINHA-VIBE and GRAPPA-VIBE for Liver MRI at 1.5 T: A Comparative In Vivo Patient Study, J Comput Assist Tomogr 2015; 39:263-269.
4. Beatty P, Brau A, Chang S, Joshi S, Michelich C, Bayram E, Nelson T, Herfkens R, Brittain J, A Method for Autocalibrating 2-D Accelerated Volumetric Parallel Imaging with Clinically Practical Reconstruction Times, In Proceedings of the 15th Annual Meeting of ISMRM, Berlin, Germany, 2007. Page 1749.
5. Robson P, Grant A, Madhuranthakam A, Lattanzi R, Sodickson D, McKenzie C, Comprehensize Quantification of Signal-to-Noise Ratio and g-Factor for Image-Based and k-Space-Based Parallel Imaging Reconstructions, Magn Reson Med 2008; 60:985-907.
Figures