1402
Patch-based super-resolution for arterial spin labeling MRI1Siemens Healthineers, Saint-Denis, France, 2Univ Rennes 1, F - 35043, Rennes, France, 3INRIA, F - 35042, Rennes, France, 4INSERM, U746, F - 35042, Rennes, France, 5CNRS, U 6074, F - 35042, Rennes, France, 6CHU Rennes, F - 35033, Rennes, France
Synopsis
Partial volume effects (PVE) are an important limitation of arterial spin labeling (ASL) acquisitions, impacting the validity of quantitative cerebral blood flow (CBF) estimations. This abstract presents a super-resolution algorithm, which includes information of high resolution (HR) structural images to reconstruct HR CBF maps from low resolution ASL series, without increasing the acquisition time. Compared with nearest neighbor, trilinear and 3rd order spline interpolations, the proposed algorithm is found to generate a CBF image closer to the one obtained with a reference HR ASL acquisition. CBF calculations can therefore be improved by using this algorithm, which reduces the PVE.
Introduction
Arterial spin labeling (ASL) provides a non-invasive quantitative evaluation of the cerebral blood flow (CBF). To acquire ASL images in a clinical compatible acquisition time, compromise must be found between signal quality and images resolution. Therefore, this modality is often acquired at low resolution, and affected by partial volume (PV) effects. Methods have been proposed to provide CBF PV correction at the voxel level.1,2 In this paper, we propose an adaptation of a patch-based super-resolution algorithm3 taking advantage of a high resolution structural image, usually mandatorily acquired in research and clinical protocols, to reconstruct CBF maps at a higher resolution, without increasing the acquisition time.Methods
Low (3.5x3.5x5mm3) and high resolution (1.75x1.75x2mm3) 2D pseudo-continuous ASL (pCASL) series with FOV=224x224mm2, TR/TE=4060/12ms, LD=1500ms, PLD=1800ms were acquired on a healthy volunteer on a 3T MR scanner. The protocol also included a 3D MP2RAGE (1x1x1mm3 isotropic, FOV=256x256mm2), using the UNI image as an unbiased structural image, and M0 images at the same low and high resolutions as equilibrium magnetization maps (TR=10s, TE=12ms).
An inhouse processing pipeline based on Nipype4, SPM8 and Python functions was used to generate perfusion images from the two raw pCASL series. The series were realigned on the first volume, the perfusion images calculated as the mean of the differences between control and label images, and the structural and M0 images were registered on these perfusion maps.
The purpose of super-resolution algorithms is to retrieve a high resolution image $$$x$$$ from a low resolution one $$$y$$$ acquired by the scanner, subject to a decimation operator $$$D$$$, a degradation model $$$H$$$, chosen as a 3D boxcar function3, and noise $$$η$$$ :
$$y = DHx + η$$
In this study, $$$x$$$ represents the high resolution perfusion or M0 image and $$$y$$$ the low resolution one. $$$\hat{x}$$$, the estimation of $$$x$$$, can be obtained by minimizing an optimization function similar to the one proposed in [3] in the context of diffusion images:
$$\hat{x} = argmin_{x}\{ || y-DHx ||^{2}_{2} + \gamma \phi_{S}(x)\}$$
where $$$γ$$$ is a scalar and $$$\phi_{S}$$$ a non-local patch-based regularization term including information from the high resolution structural image $$$x_{struct}$$$.
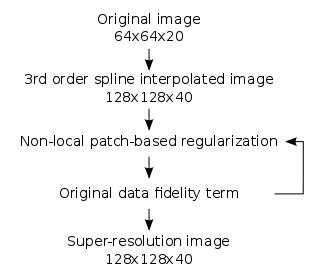
The proposed algorithm consists therefore in the following steps (figure 1). First, a 3rd order spline interpolation is performed in order to obtain an image with the desired dimensions. Then, iterations between the non-local patch-based regularization and an original data fidelity term (equations 3 and 4) until convergence generate a high resolution image minimizing the optimization problem.
$$ \hat{x}_{i}^{t+1} = \frac{1}{Z_{i}}\sum_{j\in{V_{i}}} \hat{x}_{i}^{t} exp-(\frac{||N(x_{i, struct}) - N(x_{j, struct})||^{2}_{2}}{2\sigma^{2}_{i, struct}} + \frac{||N(\hat{x}_{i}^{t}) - N(\hat{x}_{j}^{t})||^{2}_{2}}{2\sigma^{2}_{i}})$$
$$ \hat{x}^{t+1} = \hat{x}^{t+1} - (DH\hat{x}^{t+1} - y)$$
$$$N(x_{i})$$$ represents the 3x3x3 neighborhood around voxel i, $$$\sigma_{i}$$$ the empirical local variance, $$$V_{i}$$$ a 7x7x7 search volume around voxel i and $$$Z_{i}$$$ a scaling parameter controlling that the sum of the weights is equal to 1.
This algorithm was applied to the low resolution perfusion and M0 maps, using the MP2RAGE UNI image. The Buxton model was then used to calculate the CBF maps corresponding to the high resolution acquisition and the reconstructed images.5
CBF maps were also produced following nearest neighbor, trilinear and 3rd order spline interpolations applied to the perfusion and M0 maps as a matter of comparison.
Results
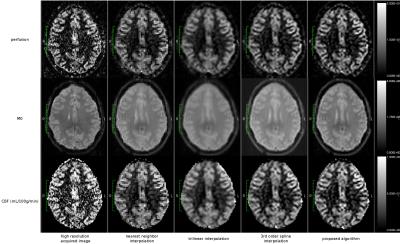
The perfusion, M0 and CBF images obtained with the different methods are presented in figure 2.
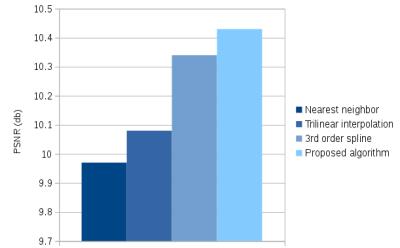
The high resolution CBF image being considered as the reference image, the quality of the reconstructions was evaluated by calculating the PSNR between this reference and the generated images (figure 3).
Discussion
The PSNR comparing the reference and the CBF map generated using the proposed algorithm is superior to the values obtained using nearest neighbor, trilinear and 3rd order spline interpolations. The contamination of the gray matter (GM) signal by white matter values induced by PV effects at low resolution and nearest neighbor interpolation, and the smoothness resulting from the trilinear and 3rd order spline interpolations, is reduced by using the proposed super-resolution algorithm. This means that the CBF values in the gray matter are closer to the one obtained with the high resolution data.
Ongoing inclusions will raise the number of subjects integrated in the results to be provided at the time of the conference.
Conclusion
The algorithm described in this paper enables the generation of high resolution CBF images from ASL series acquired at a lower resolution, without the need for an increase of the acquisition time. These images reduce the impact of PV effects in the CBF quantification, providing more reliable values, especially in GM, which is of particular interest in clinical practice.Acknowledgements
No acknowledgement found.References
1. Asllani I., Borogovac A., Brown T. R., Regression Algorithm Correcting for Partial Volume Effects in Arterial Spin Labeling MRI. Magnetic Resonance in Medicine. 2008;60:1362-1371
2. Petr J., Ferré J.C., Gauvrit J.Y., Barillot C., Denoising Arterial Spin Labeling MRI using Tissue Partial Volume. SPIE Medical Imaging. 2010, San Diego, United States, (pp. 76230L-76230L). International Society for Optics and Photonics.
3. Coupé P., Manjon J. V., Chamberland M., Descoteaux M. Hiba B., Collaborative Patch-Based Super-Resolution for Diffusion-Weighted Images. Neuroimage. 2013;83:245-261
4. Gorgolewski K., Burns C. D., Madison C., Clark D., Halchenko Y. O., Waskom M., Ghosh S. S., Nipype: a Flexible, Lightweight and Extensible Neuroimaging Data Processing Framework in Python. Frontiers in Neuroinformatics. 2011;5:13
5. Buxton R. B., Frank L. R., Wong E. C., Siewert B., Warach S., Edelman R. R., A General Kinetic Model for Quantitative Perfusion Imaging with Arterial Spin Labeling. Magnetic Resonance in Medicine. 1998;40:383-396
Figures