1314
19F imaging with physiologic motion using UTE sequence with randomized spokes orderingBijaya Thapa1,2, Kyle Jeong1,3, Insun Lee1, and Eun-Kee Jeong1,4
1Utah Center for Advanced Imaging Research, University of Utah, Salt Lake City, UT, United States, 2Department of Physics and Astronomy, University of Utah, Salt Lake City, UT, United States, 3Department of Biomedical Engeneering, Salt Lake City, UT, United States, 4Department of Radiology and Imaging Sciences, University of Utah, Salt Lake City, UT, United States
Synopsis
3D radial, such as ultra-short TE (UTE) MRI, is insensitive to motion-related ghosting artifact, mainly because of heavy over-sampling at the k-space origin. However, using the smooth view ordering, of which direction of the readout gradient smoothly changes, any small portion of FID data with motion-corruption tends to be clustered, while directions of random view ordering are spread out over the entire 3D surface. In this work, we will compare the artifact induced by both motion and missing lines in 3D UTE MRI for smooth and random view orderings.
INTRODUCTION
Hyperpolarized gases such as 3He, 129Xe etc. have been used in lung MRI over last several years to study the ventilation and perfusion maps in the lung that is useful to understand several lung diseases1. However, an expert physicist is required to produce high yield hyperpolarization and the produced gas can be used for only one time, which limits these gases for routine clinical use. As an alternative to these gases, inert fluorinated gases such as C2F6, CF4, SF6 and C3F8 have been proposed. 19F MRI may be practically useful to study the ventilation because of the following reasons: they can be used for multiple times as 19F MRI relies on thermal polarization, they are non-toxic, highly stable, and have multiple chemically equivalent fluorine atoms per molecule and generally short T1 relaxation time that enables rapid acquisition. Conventional MRI cannot be used to image gas in the lung because of short T2 due to tissue air interface. In the present study, we used 3D ultra-short TE (UTE) pulse sequence with smooth and randomized spoke ordering .The UTE sequence produces the image devoid of motion-related ghost artifact due to its heavy sampling at the k-space origin although there is position blurring due to subject’s motion along the direction of motion. Retrospective gated reconstruction (RGR), in which the measured data is sorted depending on the physiological data e.g. respiration waveform and a portion of data is discarded so that missing data spreads uniformly over the entire surface, can be used to improve the motion blurring2.METHODS
A phantom consisting a fluorinated foam on top of cardboard box containing zipper bag with a wooden block filled with C2F6 gas was placed on top of a small air-bed that is periodically inflated and deflated by a 100 CC syringe to mimic a breathing motion. The whole set up was placed inside a homemade 19F quadrature lung coil and the pressure sensor was hooked on the phantom with Velcro. The coil was matched and tuned to fluorine frequency inside the magnet. The pressure was measured using the manufacturer’s respiration belt by vertically moving the phantom to about ~1 cm through the manual pushing and pulling of the syringe and recorded into the measurement header in about every 100 ms (40th FID). The original (smooth) ordering of the radial spokes is randomly shuffled to randomize the spoke ordering. The phantom was imaged with 32,768 radial spokes, TR/TE= 2.44/0.18 ms, FOV = 288 mm and 96 readout with 192 oversampled points. The raw measurement FID data was reconstructed using our homemade image reconstruction software, which is programmed in Python and C++ languages. The FID data are sorted prior to the regrinding, based on the intensity of the physiologic data. The radial data is resampled to Cartesian grid using Kaiser-Bessel weighting3.RESULTS
Figs. 1(a-b) show the phantom mimicking the breathing motion setup and 19F lung coil. The FID of phantom obtained using radial UTE with linearly and randomly varying spokes are shown in figs. 2(a-b). The respiration signal obtained synthetically from the host computer is shown in the fig2c. and that from actual phyosiologic motion is shown in figure 2d. Figs. 3(a-d) are the images of the phantom obtained without physiologic motion. Images a and c are reconstructed with full spokes and b and d with ~50% spokes. Images a and b are acquired with linear ordering while c and d with random ordering of the spokes. Images of the phantom under exactly the same condition but with motion are shown in figs 4(a-d).DISSUSSIONS and CONCLUSIONS
From the FID plots, we see that the signal is not completely spoiled after the acquisition in linear ordering (2nd echo at the other end) while it is in randomized spoke ordering. There is clearly increased artifact with motion. With random ordering, the artifact is visibly decreased compared to linear ordering of the spokes as seen in fig.4. In the case of gas imaging, RGR, which is known to greatly reduce the artifact, does not seem help much in both linear and random ordering of the spokes. It might help in the real human lung imaging which is the main objective of the project.Acknowledgements
This work is supported by NMSS Research Grant (RG 5233- A- 2).References
[1] Chang YV et al. JMR. 2006;181:191-198.
[2] Mendes J. et al. MRM 2011;66;1268.
[3]Nielles-Vallespin S et al., MRM 2007;57;74.
Figures
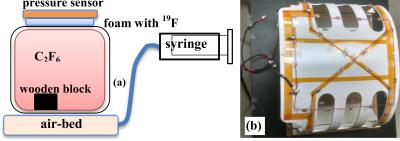
(a) schematic diagram of phantom (b) 19F
quadrature lung coil.
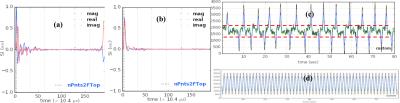
The
plot of signal intensity of FID of the 19F phantom obtained using
UTE sequence with (a) smooth and (b) randomized view orderings. Note an echo
formation at the end of the FID measured with (a) smooth ordering,
but not (b) random ordering, because the spoiler gradient after each readout
was removed to shorten the repetition time. (c) respiratory signal obtained
from physiologic motion and (d) synthetically created respiration signal from
scanner. Two red dashed lines in (c) define
FIDs data to be included in RGR.
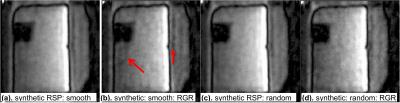
Images of phantom with synthetically generated
respiratory waveform with (a, b) smooth and (c,d) random orderings. Images
(a,c) are obtained with full data set and (b, d) with ~ 50% data set. Note that
an increased artifact in (b), which is indicated by red arrow, in smooth view
ordering with RGR. Because there is not actual motion, this artifact may be
induced by clustered missing lines during RGR. This artifact is less prominent in randomly ordered data
in (d).

Images
of phantom with physiologic motion with (a, b) smooth and (c,d) random orderings.
Images (a,c) are obtained with full data and (b, d) retrospectively gated data set
with ~ 60% of the full data. The red arrow indicates little bit reduced
artifact with RGR.