1280
Subject-specific 3D Modeling of Macaque Brain via Automatic Tissue Registration Based on in vivo MR Images Acquired at 7T1Interdisciplinary Institute of Neuroscience and Technology, Zhejiang University, Hangzhou, People's Republic of China, 2College of Biomedical Engineering & Instrument Science, Zhejiang University, Hangzhou, People's Republic of China, 3Medical Imaging Department, Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences, Suzhou, People's Republic of China, 4MR Collaboration Northeast Asia, Siemens Healthcare, Shanghai, People's Republic of China
Synopsis
Accurate subject-specific 3D modeling of macaque brain with anatomical subdivisions is important for neuroscience, neurophysiology and engineering researches. In this study, we have proposed a feasible approach for automatically creating 3D models of macaque brain based on in vivo MR images. A 3D template of macaque brain, consisting of scalp, skull, grey matter, white matter, and cerebrospinal fluid, was firstly constructed from 7T T1w images over an anesthetized macaque; then, by implementing symmetric feature-based pairwise registration method, this template was used to register another in vivo 7T dataset of macaque brain, which enables automatic and subject-specific 3D macaque brain modeling.
Purpose
Accurate subject-specific3D modeling of non-human primate’s brain with anatomical subdivisions is important for neuroscience and neurophysiology studies [1]; in addition, such structural models are needed in engineering researches such as RF coil design for high-quality MRI acquisition. Up to now, most 3D realistic-geometry models use MRI images obtained at or below 4.7T field strength. Compared to what ultra-high-field MRI can possibly enrich us, the existing models may have limited spatial resolution and hereby lose detailed anatomical information during modeling process. Moreover, creating a reliable 3D subject-specific model of, e.g., macaque brain, is complicated and workload-intensive due to manual yet cumbersome tissue segmentation. In this study, we have proposed a feasible approach for automatically creating 3D models of macaque brain based on in vivo MRI images obtained at 7T. A 3D template was firstly constructed from in vivo T1w images over a macaque, by manually segmenting the head tissues into scalp, skull, grey matter (GM), white matter (WM), and cerebrospinal fluid (CSF); Secondly, by implementing the symmetric feature-based pairwise registration method [2,3], we registered such template to another in vivo MRI dataset of macaque brain, enabling automatic and subject-specific 3D macaque brain modeling.Methods
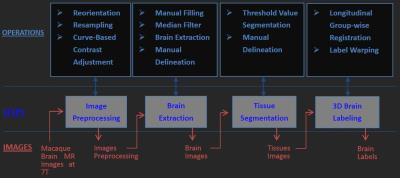
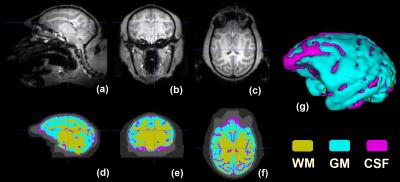
Two anesthetized macaques (case 1: male, 5.11kg, 4 year old; case 2: male, 5.2kg, 4 year old) were imaged by a Siemens MAGNETOM 7T whole-body scanner (Erlangen, Germany) with a QED knee coil (Ohio, USA). T1w images were obtained by using a 3D MPRAGE sequence (case1: TR/TE =3500/3.38 ms, GRAPPA=3, FoV=96x128 mm², 112 slices, 1mm isotropic resolution, average=10; case2: TR/TE =3500/3.38 ms, GRAPPA=3, FoV=256x200 mm², 176 slices, 0.5mm isotropic resolution, average=4). The image processing flowchart is shown in Fig. 1. It includes four major steps (as shown by four grey boxes in the middle row of Fig. 1). The original data underwent a series of pre-processing steps, including removal of non-brain slices, alignment of the brain to the center of image, reorienting and resampling of each image to a standard format (RAS coordinate, voxel size 0.5 mm isotropic, and volume size 256x256x256). To obtain an accurate template of macaque brain, all images obtained over the 1st monkey (case 1) were manually labeled into 3 tissues: scalp, skull, and cortical regions, with the aid of ITK-SNAP software [4]. Its cortical region was subsequently divided into GM, WM and CSF by implementing a threshold-value algorithm plus manual segmentation. A 3D template was then developed by merging the delineated 5 labels. Then, we registered the template to the MRI images of the 2nd macaque (case 2) by 1) affinely registration, 2) histogram matching, 3) symmetric feature-based registration, and 4) performing deformation to obtain the 3D model [2,3]. A desktop running Linux operation system (Ubuntu 14.04) with Intel i7-6700 and 32GB RAM was used, and the total computation time for the entire registration process was ~40 minutes.Results
In Figure 2, the original MRI images of the 1st monkey (case 1) and the corresponding template constructed mainly by manual segmentation are shown. The original MRI images of the 2nd monkey (case 2) and the reconstructed 3D brain model by automatic tissue registration are shown in Figure 3. In both figures, the 3D views of segmented brain tissues are illustrated for each monkey subject. Compared to the original MRI structural images, reconstructed 3D brain model through automatic tissue segmentation clearly depicts of the contour of GM, WM and CSF (Figure 3), which demonstrates the feasibility of the proposed approach.Discussion
More in vivo macaque T1w images will be acquired at 7T in future, for the purpose of evaluating and improving the proposed approach; in the meantime, to enrich our library for tissue registration, more 3D templates of macaque brain will be generated over various macaque MRI structural images and with different imaging parameters, e.g., voxel size, head position, contrast mechanism, etc, in order to consolidate the reliability of reconstructed 3D macaque brain model for each scanned monkey.Conclusion
Through manual brain tissue segmentation based on in vivo macaque T1w images acquired at 7T, a 3D structural template consisting of scalp, skull, GM, WM, and CSF has been constructed. Based on this template, a feasible approach has been proposed for automatic brain tissue registration. Its performance has been evaluated over a 2nd set of in vivo macaque T1w images acquired at 7T with high spatial resolution. It is believed that such approach can lead to subject-specific and efficient 3D modeling of macaque brain; therefore, it holds potential to largely benefit researches in neuroscience, neurophysiology, as well as engineering topics such as RF coil design in MRI.Acknowledgements
This work was supported in part by National Natural Science Foundation of China (31471052), Fundamental Research Funds for the Central Universities (2015QN81007, 2016QN81018), and Zhejiang Provincial Natural Science Foundation of China (LR15C090001).References
1. Reveley, Colin, Audrunas Gruslys, Q. Ye Frank, Daniel Glen, Jason Samaha, Brian E. Russ, Ziad Saad, Anil K. Seth, David A. Leopold, and Kadharbatcha S. Saleem. "Three-dimensional digital template atlas of the macaque brain." Cerebral Cortex (2016).
2. Dai, Yakang, Yaping Wang, Li Wang, Guorong Wu, Feng Shi, Dinggang Shen, and Alzheimer’s Disease Neuroimaging Initiative. "aBEAT: a toolbox for consistent analysis of longitudinal adult brain MRI." PloS one 8, no. 4 (2013): e60344.
3. Dai, Yakang, Feng Shi, Li Wang, Guorong Wu, and Dinggang Shen. "iBEAT: a toolbox for infant brain magnetic resonance image processing." Neuroinformatics 11, no. 2 (2013): 211-225.
4. Yushkevich, Paul A., Joseph Piven, Heather Cody Hazlett, Rachel Gimpel Smith, Sean Ho, James C. Gee, and Guido Gerig. "User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability." Neuroimage 31, no. 3 (2006): 1116-1128.
Figures