1277
Free breathing & ungated cardiac cine MRI using joint smoothness regularization on image and patch manifolds (j-STORM)1Electrical and Computer Engineering, University of Iowa, Iowa City, IA, United States, 2MR R&D, Siemens Healthcare, United States
Synopsis
A joint manifold smoothness regularization scheme (j-STORM) is proposed for free-breathing and ungated cardiac CINE imaging. The proposed method assumes that the images and square shaped image patches of the dynamic dataset live on a smooth, but separate, low-dimensional manifolds. We compare the reconstruction of two datasets from their highly undersampled measurements with the proposed j-STORM method to the image manifold smoothness scheme (STORM). The proposed scheme considerably reduces the streaking-artifacts present when only image manifold smoothness regularization is used and not the patch manifold smoothness.
Purpose
CINE imaging, which is used for functional imaging of the heart, is an integral part of most cardiac exams. Unfortunately, the need for breath-holds to freeze respiratory motion often makes it difficult to image several patient populations (e.g pediatrics, obese subjects, and patients with compromised pulmonary function.).Further, multiple breath-holds with pauses considerably increases scan-time which severly limits patient througput. The diagnostic image quality also often suffers in the absence of good breath-hold capability. Moreover, single-shot real-time imaging, which allows for cine imaging under free-breathing, suffers from limited spatial and/or temporal resolution. Thus, there is an urgent clinical need for a free breathing and ungated cardiac CINE protocol, which provides diagnostic image quality and reduces patient setup time.
Methods
We introduce a joint manifold smoothness regularization scheme, which assumes that the images ($$$\mathbf x_1, ... , \mathbf x_n$$$) and square shaped image patches $$$\{P_r(\mathbf X); \mathbf r \in \Omega\}$$$ live on a smooth, but separate, low-dimensional manifolds, to regularize the reconstruction of the highly undersampled dynamic dataset. This work builds upon the image manifold smoothness regularization1 scheme (STORM), where the similarity of images at the same cardiac/respiratory phases were exploited. The recovery was aided by a modified golden angle acquisition, where 3-4 kspace lines are repeated in every image time frame
$$ \underbrace{\begin{bmatrix}\mathbf z_i\\ \mathbf x_i\end{bmatrix} }_{\mathbf y_i}= \underbrace{\begin{bmatrix}\mathbf \Phi\\ \mathbf B_i\end{bmatrix} }_{\mathbf A_i} \mathbf x_i + \mathbf n_i.$$
Here $$$\mathbf \Phi$$$ denotes the common measurements, while $$$\mathbf B_i$$$ denotes the measurements that are different from frame to frame. The structure of the image manifold, or equivalently the derivative (Laplace Beltrami) operator denoted by $$$\mathbf Q$$$, was estimated from navigator data $$$\mathbf Z = \mathbf \Phi\mathbf X$$$. While the image manifold regularization scheme exploited the non-local similarities between images in the time series, it fails to capture the redundancies within each image as well as between adjacent images. Motivated by our patch based implicit motion compensated reconstruction scheme2, we add a patch manifold prior to further constrain the problem:
$$\mathbf X^* = \arg\min_{\mathbf X} \left\lbrace \|\mathcal{A}(\mathbf X) - \mathbf b\|_F^2 + \lambda_1\|\mathbf X\mathbf Q\|_2^2 + \lambda_2 ~\varphi(\mathbf X) \right\rbrace.$$
Here, $$$\mathcal{A}$$$ is the collective measurement operator and $$$\lambda_i$$$ are the regularization parameters. The function $$$\varphi$$$ can be chosen as any penalty function that penalizes the distance between the extracted patches in the low-dimensional manifold. In this work, we consider a VBM3D3 prior; the VBM3D algorithm was originally introduced to denoise video sequences using sparse 3D transform domain collaborative filtering of extracted image patches. We solve the optimization problem using the proximal forward backward splitting (PFB) algorithm4, which consists of two steps:
1. Solve the quadratic problem consisting of the first two terms in the proposed objective function using the conjugate gradient method.
2. Solve the patch denoising step: $$$\mathbf X^* = \arg\min_{\hat{\mathbf X}}\|\tilde{\mathbf Y} - \hat{\mathbf X}\|_F^2 + \varphi(\hat{\mathbf X})$$$. Here we use the standard VBM3D solver3.
We consider two datasets in this study. The first dataset was acquired on a 3T scanner (Siemens Skyra) using a prototype FLASH sequence (TR/TE 4.3/1.92 ms, FOV=300mm, Base resolution=256, 10000 radial spokes, scan time=43 s. Each frame had 10 radial lines out of which 4 were navigator lines, resulting in a temporal resolution of around 43 ms. The second dataset was acquired using a SSFP sequence on a Siemens 3T TIM Trio scanner with a 18 channel cardiac array (TR/TE = 4.2/2.2 ms, slices = 5, slice thickness = 5 mm, FOV = 300mm, spatial resolution = 1.17 mm). A temporal resolution of 42ms was achieved by sampling 10 lines of k-space per frame, out of which 4 were navigator lines. 10000 radial lines of kspace were acquired per slice which resulted in an acquisition time of around 42 s per slice.
Results
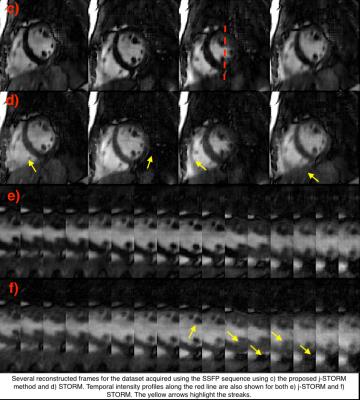
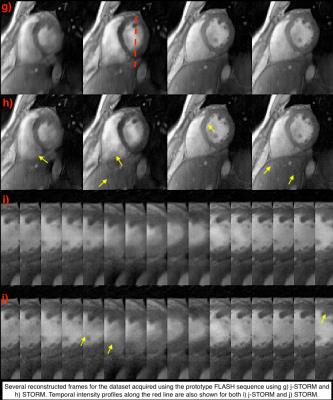
We compare the proposed j-STORM method with the state-of-the-art STORM method for the two datsets described above. Figure (c) shows the reconstructed frames from the second dataset using the j-STORM method Figure (d) shows the reconstruction using STORM. As highlighted by the yellow areas, the streaking artifacts present in reconstructed images using STORM are considerably attenuated using the proposed j-STORM method. Similar behavior is seen for the dataset acquired using the prototype FLASH sequence (see Fig. (g)-(h)).Conclusion
We proposed a novel free-breathing cardiac dynamic imaging method using a joint image and patch manifold prior. We assume that the images and the non-local patches in the dynamic dataset live in a smooth low-dimensional but separate manifold. The proposed method not only reduces streaking-artifacts that are present when using image manifold smoothness only, but also improves image quality.Acknowledgements
No acknowledgement found.References
1. S. Poddar and M. Jacob, Dynamic MRI using Smoothness Regularization on Manifolds (SToRM), IEEE Transactions on Medical Imaging, 35(4), April 2016.
2. Y. Mohsin, S.G Lingala, E. DiBella, and M.Jacob, Accelerated dynamic MRI Using Patch Regularization for Implicit motion CompEnsation (PRICE), Magnetic Resonance in Medicine, in press.
3. K. Dabov, A. Foi, and K. Egiazarian, Video denoising by sparse 3D transform-domain collaborative filtering, Proc. 15th European Signal Processing Conference (EUSIPCO), Sep. 2007.
4. P. L. Combettes and V. R. Wajs, Signal Recovery by Proximal Forward-Backward Splitting, Multiscale Model. Simul., 4(4), 1168–1200, Jul. 2006.
Figures