1252
Accelerated Magnetic Resonance Spectroscopic Imaging Using Readout Segmentation (ASPIRES)1Fraunhofer MEVIS, Bremen, Germany
Synopsis
ASPIRES is a novel method for accelerated, high-resolution, large-bandwidth echo-planar spectroscopic imaging. It uses readout segmentation to decouple the echo spacing from the spatial resolution and gradient-system performance. Readout segmentation is combined with blipped phase encoding to accelerate scans by random undersampling along the readout-segment, phase-encoding and time directions. An elliptical acquisition window in the readout-phase encoding plane reduces scan time further. Distributed-multisensory compressed sensing reconstruction efficiently restores signal properties in both spatial and frequency domains. ASPIRES promises to enhance the diagnostic performance of MR spectroscopic imaging in clinical routine and improve the study of metabolites at high field strengths.
Purpose
An efficient acquisition method for Magnetic Resonance Spectroscopic Imaging (MRSI)1 is Echo-Planar, Spectroscopic Imaging (EPSI)2, but this technique is severely limited by a low spectral bandwidth, particularly at high field strength, at high spatial resolution or for metabolites with a large range of chemical shifts. This paper introduces an accelerated magnetic resonance spectroscopic imaging using readout segmentation (ASPIRES), which combines a high-resolution, large-bandwidth MRSI3 with strong scan acceleration in order to meet the time constraints of clinical imaging.Methods
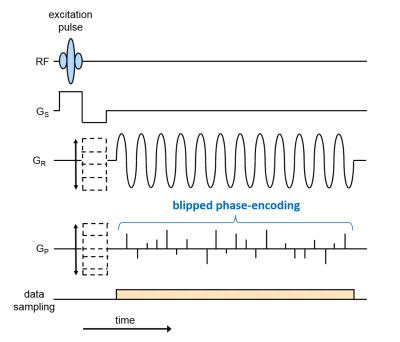
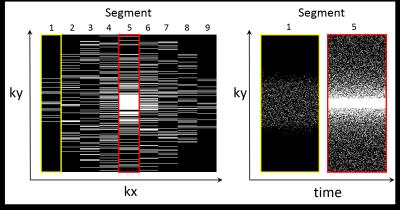
Fig. 1 shows the proposed pulse sequence diagram for readout (RO) segmentation with stepped encoding gradients in both readout (kx) and phase-encoding (ky) directions. A variable blipped phase-encoding (PE) gradient allows random undersampling that varies from echo to echo4. In the current study fully sampled data from a transverse slice through the orbits of a healthy volunteer were acquired to allow comparing results derived from retrospectively undersampled data sets. A Siemens 3T Skyra system and a 32-channel head coil were used. Imaging parameters were as follows: FOV 200mm; matrix 512 x 338; 9 RO segments; slice thickness 2mm; TR 100ms; 144 echoes with spacing 520μs, corresponding to a spectral bandwidth of 1.9kHz; scan time 5:04 mins. Single-echo and separate fat and water images were reconstructed as described previously3. The acquired data set was retrospectively randomly undersampled in three dimensions: RO segment (kx), PE (ky) and time. Elliptical view5 sampling in kx and ky were applied (see fig. 2). A distributed multisensory implementation6 of compressed sensing reconstruction7 was used, including sparsifying operators both in space and in time. The required receive-coil sensitivity maps were calculated from the undersampled data by using the adaptive combined technique8.Results
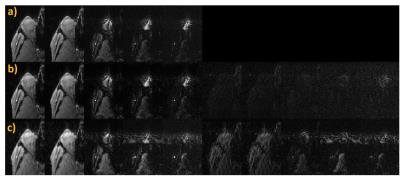
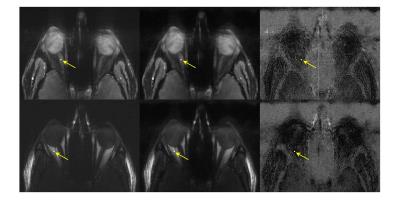
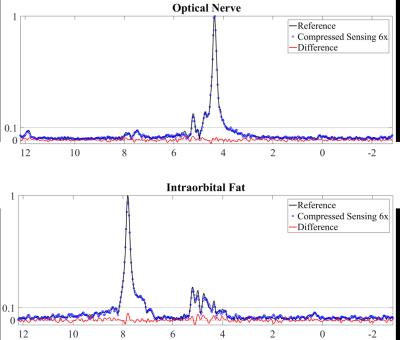
Fig. 3 shows the fully-sampled reference images together with a six-fold accelerated ASPIRES images and the corresponding zero-filled reconstruction. The ASPIRES images are visually very similar to the reference images and show considerably enhanced detail compared to a zero-filled reconstruction of the undersampled data. As shown in fig. 4, the reconstruction scheme also generated spectrally selective fat and water images9 with good preservation of the contrast and anatomical detail seen into the images derived from the fully sampled data. As seen in fig. 5, the corresponding local spectra generated from the undersampled and fully sampled data sets show very close agreement with respect to detailed spectral features.Discussion
Readout segmentation enables the combination of high spatial resolution and high spectral bandwidth, since it decouples the spatial resolution along the RO direction from the echo spacing. The drawback is a time penalty, due to the acquisition of multiple RO segments, that can be mitigated by exploiting the extra degree of freedom provided by RO segmentation when designing undersampling strategies. Therefore, RO segmentation provides two benefits: firstly it allows an elliptical acquisition window by using a different ky extent for each RO segment; secondly, it increases the randomness of the sampling scheme in the kx-ky plane by allowing ky sampling schemes that vary between RO segments. Additional sampling randomness is achieved by using a variable-PE blipped gradient to modify the ky sampling scheme for each echo. As a consequence, ASPIRES can achieve a strong scan acceleration, allowing to compensate for the time penalty due to RO segmentation. In the proposed example, the scan-time reduction was from 5 mins to 50 secs. The resulting protocol requires a 50% increase in scan time compared to a standard EPSI measurement, but provides a five-fold increase in spatial resolution for the same spectral bandwidth. Further investigations will be performed to optimize the compressed sensing regularization parameters and to exploit the increased image-domain sparsity at later echo times due to T2* decay.Conclusions
MRSI can provide important information for clinical diagnosis and treatment monitoring, but the technique is not in widespread use because the standard sequence requires long scan times that are often unsuitable for the patient examinations. EPSI reduces this scan time substantially, but is subject to gradient-hardware limitations. The proposed ASPIRES technique can provide a similar improvement in scan time, but is not restricted by hardware limitations, allowing a flexible choice of spatial resolution and spectral bandwidth. The technique has the potential to increase the clinical application of MRSI for a wide range of major diseases such as cancer and dementia. The high-bandwidth capability of the method will be of particular interest for metabolite studies at ultra-high field strengths and for measurements with nuclei other than 1H.Acknowledgements
No acknowledgement found.References
1. Posse S, Otazo R, Dager SR, Alger J. MR spectroscopic imaging: principles and recent advances. Journal of Magnetic Resonance Imaging. 2013 Jun 1;37(6):1301-25.
2. Sarkar S, Heberlein K, Metzger GJ, Zhang X, Hu X. Applications of high-resolution echoplanar spectroscopic imaging for structural imaging. Journal of Magnetic Resonance Imaging. 1999 Jul 1;10:1-7.
3. Porter DA, Vicari M. Readout Segmentation for Increased Spectral Bandwidth in High Spatial and Spectral Resolution (HiSS) MRI. Proceedings of the 24rd Annual Meeting ISMRM. 2016.
4. Hu S, Lustig M, Chen AP, Crane J, Kerr A, Kelley DA, Hurd R, Kurhanewicz J, Nelson SJ, Pauly JM, Vigneron DB. Compressed sensing for resolution enhancement of hyperpolarized 13 C flyback 3D-MRSI. Journal of magnetic resonance. 2008 Jun 30;192(2):258-64.
5. Bernstein MA, Fain SB, Riederer SJ. Effect of windowing and zero-filled reconstruction of MRI data on spatial resolution and acquisition strategy. Journal of Magnetic Resonance Imaging. 2001 Sep 1;14(3):270-80.
6. Otazo R, Kim D, Axel L, Sodickson DK. Combination of compressed sensing and parallel imaging with respiratory motion correction for highly-accelerated first-pass cardiac perfusion MRI. Magn Reson Med. 2010;64(4):1148-54.
7. Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magnetic resonance in medicine. 2007 Dec 1;58(6):1182-95.
8. Walsh DO, Gmitro AF, Marcellin MW. Adaptive reconstruction of phased array MR imagery. Magnetic Resonance in Medicine. 2000 May 1;43(5):682-90.
9. Medved M, Newstead GM, Abe H, Zamora MA, Olopade OI, Karczmar GS. High spectral and spatial resolution MRI of breast lesions: preliminary clinical experience. American Journal of Roentgenology. 2006 Jan;186(1):30-7.
Figures