1251
Comparison of Acceleration Methods for Brain MRSI at 7T1Department of Biomedical Imaging and Image-guided Therapy, High Field MR Centre, Medical University of Vienna, Vienna, Austria, 2Department of Electrical Engineering and Computer Science, MIT, Cambridge, MA, United States, 3Christian Doppler Laboratory for Clinical Molecular MR Imaging, Medical University of Vienna, Vienna, Austria
Synopsis
In this work three different acceleration methods, 2D-CAIPIRINHA with phase-encoding, spiral encoding and concentric circles (CONCEPT) for brain MRSI at 7T were compared. The metabolic maps were compared qualitatively, and the CRLB and SNR values were compared quantitatively. The metabolic maps of 2D-CAIPIRINHA with phase-encoding provided the best data with respect to metabolic maps, CRLB and SNR values. Improving the PSF by increasing the matrix size from 48x48 to 64x64 enhanced the data quality again for CONCEPT. Spiral and CONCEPT encoding are prone to spectral baseline distortions. Nevertheless, CONCEPT has a high potential for high resolution brain MRSI at 7T.
Introduction
Two major problems that prevent the widespread application of MRSI in clinical routine and neuroscience are low SNR/t and long scan times. The low SNR/t can be addressed by high magnetic fields, measuring the free induction decay (FID) directly, and using array coil.1,2 Long measurement times can then be reduced by omitting k-space encoding steps (e.g. Parallel Imaging such as 2D-CAIPIRINHA), or spatio-spectral encoding (SSE) such as spiral, EPSI or concentrically circular echo-planar trajectories (CONCEPT)3. Although CAIPIRINHA was shown to be a versatile tool for accelerating phase-encoded MRSI4, the maximum acceleration, R, is limited to R~10, which is not enough for 3D-MRSI. Spiral encoding was shown to provide good results at 3T5, but has a lower SNR/t-efficiency at 7T, which decreases with increasing matrix size and spectral bandwidth. CONCEPT has a non-uniform k-space density, but if the target k-space density is a Hamming filter it can provide high efficiencies even at 7T with high resolutions, as it is self-rewinding. This work compares 2D-CAIPIRINHA and two SSE acceleration methods (i.e., spiral and CONCEPT) for brain MRSI at 7T.Methods
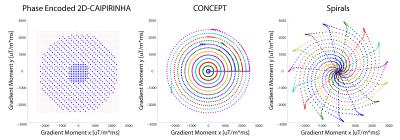
All brain MRSI scans were performed at 7T with a 32-channel array coil using three 2D-acceleration approaches: 2D-CAIPIRINHA, constant-density spiral, and CONCEPT encoding (see Figure 1). The data were density compensated and Hamming weighted in post-processing. All three sequences measured the FID with an acquisition delay of 1.3 ms, a TR of 600 ms, a FoV of 200x200 mm, and a 12 mm slice thickness.2 Prescans for estimating the coil combination weights for the MUSICAL method6 were included in all sequences.The sequence parameters were as follows:
2D-CAIPIRINHA with phase-encoding: 48x48 matrix size, elliptically encoded, 2D-CAIPIRINHA with R = 3, 272 ms acquisition window, 6000 Hz spectral bandwidth and 1632 FID-points (cut in post-processing to match CONCEPT), total measurement time 6m 20s.
Spiral: 48x48 matrix size, 272 ms acquisition window, 1852 Hz spectral bandwidth and 512 FID-points (cut in post-processing), two temporal interleaves (TI), total measurement time 6m 40s.
CONCEPT: 64x64 matrix size, cut down to 48x48 in post-processing (and also left at 64x64 for comparison), 272 ms acquisition window, 1852 Hz spectral bandwidth 512 FID points, two TIs, total measurement time 8m 20s (64x64) and 6m 20s (48x48).
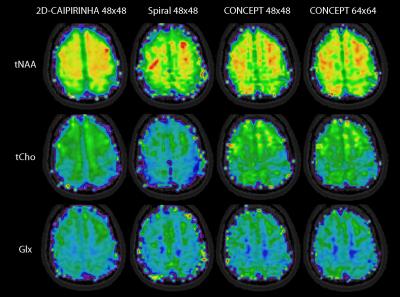
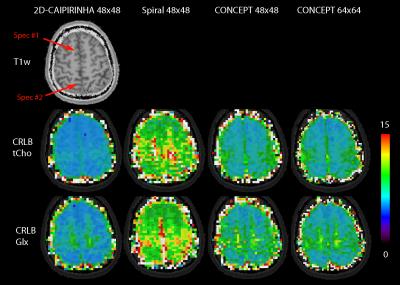
All spectra were fitted with LCModel. Metabolic, SNR, and CRLB maps of major metabolites (Glx, tNAA, and tCho) were compared qualitatively and quantitatively between 2D-CAIPIRINHA, spiral, and CONCEPT (both 48x48 and 64x64).
Results
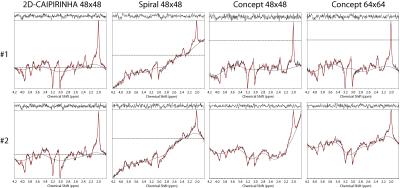
Metabolic maps are shown in figure 2, and the corresponding CRLB maps in figure 3. Red arrows show the location of two spectra which are provided in figure 4. The baseline of the spiral and CONCEPT spectra were often curved, thereby complicating reliable LCModel quantification. Spectra #2 show an example where the higher resolution of the CONCEPT 64x64 improved the spectral quality by reducing the lipid contamination. The SNR (median ± median absolute deviation) as reported by LCModel for 2D-CAIPIRINHA, spiral, CONCEPT 48x48, and CONCEPT 64x64 were 16.0±1.8, 9.0±1.6, 13.0±2.3, 13.0±2.8, and the Glx CRLB values were 5.0±1.0, 9.0±2.0, 6.0±1.7 and 6.0±1.8, respectively.Discussion
Due to the high bandwidth and high resolution demands, 42% of the spiral trajectory consisted of rewinders, reducing its sampling efficiency to sqrt(1-0.42) ~ 76%, which is about the observed SNR ratio to CONCEPT of 9/13 ~ 69%. CONCEPT provides intrinsically closed trajectories, thus having 100% sampling efficiency. However, it is important to stress that spiral, EPSI or other trajectories have sampling efficiencies of ≥95% at lower fields (≤3T) or lower resolutions (≤32x32). CONCEPT has a sampling density of 1/k, where k is the k-space radius. If a sinc PSF is requested, the density efficiency of CONCEPT is 87 %. However, if the data are anyway Hamming filtered during post-processing, the density efficiency of CONCEPT is over 100% in comparison to constant density phase-encoded sampling.
Another advantage of CONCEPT is that higher resolutions are possible at 7T than with spirals, which seem to improve data quality as indicated by the Glx map and spectrum #2. Resolutions of 64x64 with a 200x200 mm² are not possible for spirals with two TIs.
In this preliminary comparison the data quality of the three compared SSE methods is lower than for the phase-encoded sequence, because residual artifacts complicated the fitting process. This will be addressed in future studies. Nevertheless, this work shows that SSE sequences, and in particular CONCEPT, have a high potential for high spatial resolution MRSI at 7T, especially if 3D encoding is targeted.
Acknowledgements
This study was supported by the Austrian Science Fund (KLI-61) and FFG Bridge Early Stage Grant No. 846505.References
1: Bogner W, Gruber S, Trattnig S, et al. High-resolution mapping of human brain metabolites by free induction decay (1)H MRSI at 7 T. NMR Biomed. 2012 Jun;25(6):873-82.
2: Henning A, Fuchs A, Murdoch JB, Boesiger P. Slice-selective FID acquisition, localized by outer volume suppression (FIDLOVS) for (1)H-MRSI of the human brain at 7 T with minimal signal loss. NMR Biomed. 2009 Aug;22(7):683-96.
3: Furuyama JK, Wilson NE, Thomas MA. Spectroscopic imaging using concentrically circular echo-planar trajectories in vivo. Magn Reson Med. 2012 Jun;67(6):1515-22.
4: Strasser B1, Považan M, Hangel G, et al. (2 + 1)D-CAIPIRINHA accelerated MR spectroscopic imaging of the brain at 7T. Magn Reson Med. 2016 Aug 22 [Epub].
5: Andronesi OC, Gagoski BA, Sorensen AG. Neurologic 3D MR spectroscopic imaging with low-power adiabatic pulses and fast spiral acquisition. Radiology. 2012 Feb;262(2):647-61.
6: Strasser B, Chmelik M, Robinson SD et al. Coil combination of multichannel MRSI data at 7 T: MUSICAL. NMR Biomed. 2013 Dec;26(12):1796-805.
Figures