1231
Realistic 4D abdominal phantom for magnetic resonance imaging1Dept. of Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States, 2Dept. of Radiology, University Hospitals of Cleveland, Cleveland, OH, United States
Synopsis
Validation and evaluation of novel data acquisition and reconstruction strategies are major challenges in abdominal magnetic resonance imaging, and particularly in quantitative imaging. Here, a new 4D numerical abdominal phantom combining anatomical morphology, respiratory motion, tissue properties, and physiological function is introduced to enable comparison of different data collection and reconstruction schemes for abdominal MRI.
Purpose
The variety of newly introduced techniques for 3D abdominal imaging and quantitative mapping has led to an increased need for validation, evaluation, and comparison of these novel image acquisition and reconstruction strategies1. However, validation and comparison of such techniques is complicated due to the lack of reference data (“ground truth”); the need to compensate for respiratory motion and the desire for high resolution large coverage generally preclude the use of in vivo reference images. Moreover, most of the existing abdominal digital phantoms are only capable of simulating either anatomical morphology, respiratory motion, tissue properties, or physiological function2. Here, a 4D numerical abdominal phantom is presented in which tissue properties (T1 and T2), perfusion, fat fraction, diffusion, and respiratory motion are included. This realistic abdominal MR phantom can be used to simulate different contrast preparations and sampling schemes under controlled conditions, enabling comparisons between various reconstruction techniques in a way that is not possible with in vivo data.Methods
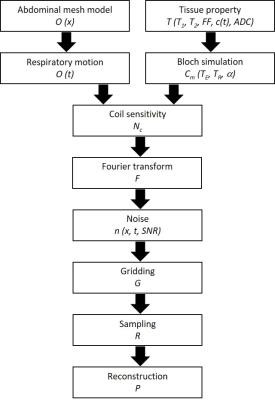
The numerical abdominal phantom was based on the in-vivo human CT abdomen polygon meshes of National Alliance for Medical Image Computing (NAMIC)3 and then voxelized to generate anatomical models ($$$O(\overrightarrow{x})$$$) for different tissues, including vessel, bone, intestine, gallbladder, adrenal gland, muscle, kidney, ureter, liver, pancreas, spleen, stomach, and fat1. Simulated lesions can be added with user-specified shapes, sizes and locations. Mathematically, the abdominal phantom is described in k-space ($$$D(\overrightarrow{k},t)$$$) through a combination of several weighting functions: $$D(\overrightarrow{k},t)= P⋅R⋅G⋅[F⋅N_c⋅C_m (T_E,T_R,α)⋅T(T_1,T_2,FF,c(t),ADC)⋅O(\overrightarrow{x},t)+n(\overrightarrow{x},t,SNR)].$$ A non-rigid deformation model for respiratory motion ($$$O(t)$$$) was created from MR scans of a healthy volunteer. Different tissue properties (T1 and T2) are specified for each tissue and signal models for physiological function (fat fraction: FF, dynamic contrast concentration: c(t), and diffusivity: ADC) are specified for liver (Fig.1). Two identical phantoms with different chemical–shift frequencies for water and a single-peak fat (440Hz at 3T) were generated using Bloch simulation with scanning parameters (Cm: TE, TR and α). The water and fat phantom were then combined to obtain signals with varying levels of fat fraction in the liver4. For the dynamic contrast enhancement signal model, a dual-input single-compartment model was used to retrieve liver perfusion parameters from dynamic contrast-enhanced MRI data5. The dynamic contrast concentration was used to simulate the enhancement pattern of signal in the liver. Tissue diffusivity with different b-values were used to create diffusion-weighted images6-7 and multiplied to previous signal. After simulating tissue properties, fat fraction, dynamic contrast and diffusivity of each voxel, the phantom images were multiplied by actual coil sensitivity maps (Nc) from MR scans to create multi-coil images and Fourier transformed (F) to k-space. Gaussian noise was added in k-space to achieve the desired SNR in the simulated source data ($$$n(\overrightarrow{x},t,SNR)$$$). Arbitrary trajectories (G) and undersampled acquisitions (R) can be simulated by specifying the desired sampling trajectory and reconstruction methods (P).Results
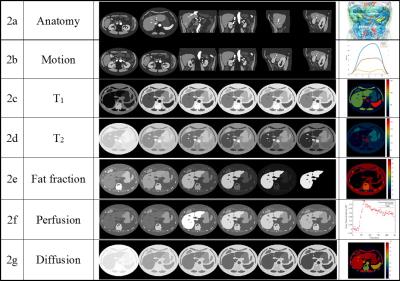
Figure 2 shows representative images depicting different respiratory states and contrasts that are possible using this digital phantom. The images were reformatted to axial, coronal, sagittal plane at different position (2a). The effects of non-rigid respiratory motion along three directions (2b) can be seen by comparing the images in the first row and the second row. The third row shows a slice undergoing T1 signal recovery after an inversion recovery pulse (2c). Similar to T1 simulation, T2-weighted images after T2 preparation can be simulated (2d). Water-only images are shown in the fifth row, where the fat percentage in the liver has been varied (0% fat, 20%, 40%, 60%, 80% and 100%) (2e). Dynamic contrast enhanced images can be simulated at pre-contrast, 20 sec, 70 sec, 120 sec, 180 sec and 240 sec post-contrast in the liver (2f). Diffusion weighted images can be generated using different b-values (0, 200, 400, 600, 800 and 1000 s/mm2) (2g).Discussion
In this work, a 4D numerical abdominal phantom has been proposed to enable realistic and flexible parameter simulation to validate image acquisition, reconstruction and quantitative approaches. Example phantom images demonstrate the feasibility of simulated anatomical morphology, respiratory motion, tissue properties, and physiological function using the presented framework. Moreover, a key advantage of quantification using this abdominal phantom is that noiseless ground truth is always available for assessment of errors. There is no partial volume effect in the phantom, but this can be achieved by adding a low-pass filter module.Conclusion
The proposed 4D numerical abdominal phantom enables versatile and realistic simulations of abdominal MR with respiratory motion.Acknowledgements
Siemens Healthcare, R01DK098503, R01HL094557.References
1. Chen Y, Jiang Y, Pahwa S, et al. MR Fingerprinting for Rapid Quantitative Abdominal Imaging. Radiology. 2016 Apr;279(1):278-86.
2. Segars WP, Sturgeon G, Mendonca S, et al. 4D XCAT phantom for multimodality imaging research. Med Phys. 2010 Sep;37(9):4902-15.
3. http://hdl.handle.net/1926/543 National Alliance for Medical Image Computing: CT Abdomen. Copyright (c) 2011, Canadian Light Source, developed by Elise Norm and All rights reserved.
4. Tsao J and Jiang Y. Hierarchical IDEAL: Fast, Robust, and Multiresolution Separation of Multiple Chemical Species from Multiple Echo Times. Magn Reson Med. 2013 Jul;70(1):155-9.
5. Chen Y, Lee G, Wright K, et al. Free-breathing liver perfusion imaging using 3-dimensional through-time spiral generalized autocalibrating partially parallel acquisition acceleration. Invest Radiol. 2015 Jun;50(6):367-75.
6. Kaya B and Koc Z. Diffusion-weighted MRI and optimal b-value for characterization of liver lesions. Acta Radiol. 2014 Jun;55(5):532-42.
7. Morani A, Elsayes K, Liu P, et al. Abdominal applications of diffusion-weighted magnetic resonance imaging: Where do we stand. World J Radiol. 2013 Mar 28; 5(3): 68–80.
Figures