1177
Image-registered whole mount histology technique for validation of MRgFUS therapies1Radiology and Imaging Sciences, University of Utah, Salt Lake City, UT, United States, 2Bioengineering, University of Utah, Salt Lake City, UT, United States, 3Pathology, University of Utah, Salt Lake City, UT, United States, 4Surgery, University of Utah, Salt Lake City, UT, United States
Synopsis
Clinical pathological review of histological samples is the gold standard for identification of tissue damage. For the development of new magnetic resonance-guided focused ultrasound (MRgFUS) procedures, in vivo post-treatment imaging assessment techniques require correlation to histopathology. Our study presents methodology and preliminary preclinical results of a new MRI-registered whole mount histology technique for efficacy evaluation of MRgFUS therapies.
Introduction
MRgFUS is a non-invasive technology that delivers focused energy deep inside the body. Although not a new idea, MRgFUS is a novel, unproven intervention for several important indications, including breast and other cancers. Before MRgFUS can be offered as a completely non-invasive treatment option for malignant disease, treat-and-resect validation studies are needed to conclusively demonstrate the efficacy of the MRgFUS ablation procedure. To enable these pivotal efficacy trials, methods that provide timely accurate assessment of treatment efficacy must be developed and validated.
For interventional procedures, post-treatment imaging techniques are validated through correlation studies of the in vivo imaging results to histopathology. This is a challenging problem as once anatomic orientation to the body is lost, it is difficult to reorient the excised specimen. Several techniques of varying accuracy have been developed to address this problem1-5 with the goal of retaining more anatomic information from the tissue sample for improved registration and analysis. Our study complements this work with a new MRI-registered whole mount histology technique for the evaluation of MRgFUS therapy.
Methods
Animal Models: All experiments were approved by the local animal committee. Tumor cell suspensions were generated by disassociating VX2 tumors in collagenase and prepared in a 50:50 mixture of media (~1 x106 cells) and Matrigel. The suspension was injected bilaterally into the cranial thigh muscles of White New Zealand rabbits (N=4). After 14 days, a biopsy marker (18g HydroMARK Biopsy Site Marker) was placed within the central portion of one of the tumors in each animal under ultrasound guidance.
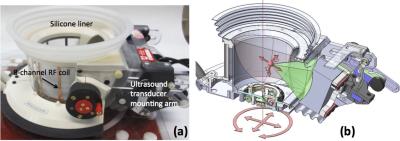
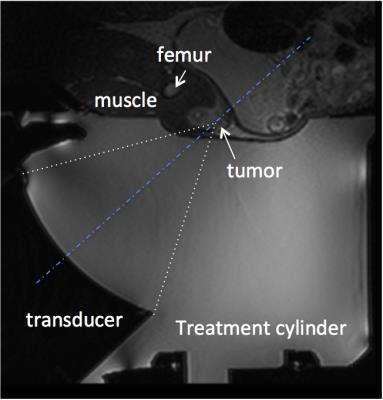
MRgFUS ablation procedure: All MRgFUS ablation procedures were performed with a breast-specific MRgFUS system6 inside a Siemens Trio 3T MRI scanner. The MRgFUS system (shown in Fig. 1) has an ultrasound phased array transducer laterally mounted on a treatment cylinder, providing numerous degrees of freedom for transducer positioning. Four days after the biopsy markers were placed, the anesthetized rabbits underwent MRgFUS partial tumor ablation. A T2-weighted sagittal image of the rabbit placed on the MRgFUS system is shown in Figure 2. Each rabbit received 4-8 individual sonications (~40 W, 40 second) targeting the tumor. Ablations were monitored by MR thermometry. Contrast enhanced MR images (CE-MRI) using a gadolinium agent (MultiHance, 0.01 mmol/kg) were obtained approximately 30 minutes after ablation. Seven days post ablation, animals were sacrificed and the tumors were excised.
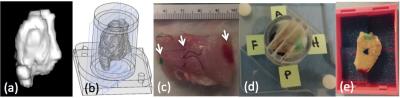
Whole mount histology procedure: Each step of the excision and mounting procedure is seen in Figure 3. The volume of tissue to be excised was segmented (Osirix Lite v8.0.1) from 3D T1w MR images and imported into a solid modeling CAD program (Solidworks) from which a custom tissue holder was designed that allowed the tumor to be oriented and sliced along the MRI acquisition planes. After excision, the tissue was anatomically oriented to the in vivo orthogonal MRI planes through marker placement and then placed in the tissue holder. Using a custom multi-blade tool, 5mm thickness slices were placed in MRI-oriented large tissue-embedding cassettes for standard processing and later stained with hematoxylin and eosin (H&E). Tumor dimensions in the whole mount H&E histology slides were compared to T1w CE-MRI with a paired t-test.
Results
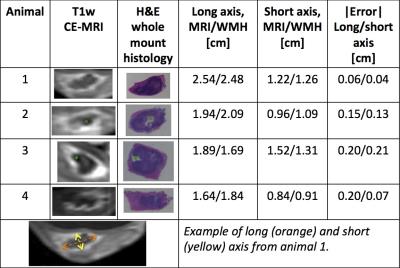
Partial tumor ablation was successful with accumulated thermal dose measured during the MRgFUS sonications qualitatively agreeing with the non-enhancing CE-MRI images obtained post-ablation. A comparison of these CE-MRI images and the H&E whole mount histology prepared after tissue excision seven days after ablation is shown in Figure 4. The shape of the hyperintense tumor rim on CE-MRI and the tumor (purple) embedded in muscle tissue (pink) on the H&E slides quantitatively agree (p < 0.05). In animals 2 and 3, the position of the biopsy clip can be seen on both the CE-MRI and H&E slides. A comparison of the long and short axis of the tumor in both image formats gives an error range of 4-13%.Discussion & Conclusion
While non-invasive MRgFUS procedures hold significant potential, acute assessment of tissue viability after the ablation procedure is critical, particularly for malignant tissues. The development and evaluation of techniques that correlate MR images to histopathological data is critical to answer this question. This treat-and-resect study demonstrates the potential of registering histopathological data to MR images. While this preliminary data shows quantitative agreement between CE-MRI and MRI-registered H&E data, the sample size and delay between imaging and tissue harvest limits the results. The continued development and evaluation of MRI-registered histology techniques will allow rigorous evaluation of MRgFUS procedures providing more treatment options for patients.Acknowledgements
This work was funded by NIH R01 CA172787.References
1. Clarke, G. M. et al. Whole-specimen histopathology: a method to produce whole-mount breast serial sections for 3-D digital histopathology imaging. Histopathology 50, 232-242, (2007).
2. Langer, D. L. et al. Prostate cancer detection with multi-parametric MRI: logistic regression analysis of quantitative T2, diffusion-weighted imaging, and dynamic contrast-enhanced MRI. J Magn Reson Imaging 30, 327-334, (2009).
3. Shah, V. et al. A method for correlating in vivo prostate magnetic resonance imaging and histopathology using individualized magnetic resonance-based molds. Rev Sci Instrum 80, 104301, (2009). 4. Clarke, G. M. et al. Whole-mount pathology of breast lumpectomy specimens improves detection of tumour margins and focality. Histopathology 69, 35-44, (2016).
5. Clarke, G. M. et al. 3D Pathology Volumetric Technique: A Method for Calculating Breast Tumour Volume from Whole-Mount Serial Section Images. Int J Breast Cancer 2012, 691205, (2012).
6. Payne, A. et al. Design and characterization of a laterally mounted phased-array transducer breast-specific MRgHIFU device with integrated 11-channel receiver array. Med Phys 39, 1552-1560, (2012).
Figures