1155
Chemogenetic fMRI and 18F-FDG PET Reveal Functional Projections of Hoxb1-Derived Noradrenargic Neurons1Biomedical Research Imaging Center, Department of Neurology, University of North Carolina Chapel Hill, Chapel Hill, NC, United States, 2Developmental Neurobiology, NIEHS/NIH
Synopsis
In this study, we show that chemogenetic fMRI and 18 F FDG PET can sensitively dissect the functional neurocircuits of noradrenergic cells derived from rhombomere 4 expressing Hoxb1 during neurodevelopment. To address this, we used a novel genetically engineered mouse line expressing Designer Receptors Exclusively Activated for Designer Drugs (DREADD) in Hoxb1-derived noradrenargic neurons in several subpopulations throughout the pons and medulla. For the first time, we used ICA-based fMRI analaysis to dissect the complex polysynaptic pathways associated with chemogenetic modulation of Hoxb-1 derived noradrenargic neurons.
Purpose
Beyond their heterogeneity with regards to anatomical location, firing patterns and function, many of the noradrenargic neurons that populate the brainstem are derived from genetically similar progenitors. To date, limited techniques are available to noninvasively dissect the functional role of these genetically similar subpopulations. This study investigates whether chemogenetic fMRI and 18F-FDG PET can sufficiently isolate the functional neurocircuits of noradrenergic cells derived from rhombomere 4 which express Hoxb1 during neurodevelopment. To address this, we used a novel genetically engineered mouse line expressing Designer Receptors Exclusively Activated for Designer Drugs (DREADDs) in Hoxb1-derived noradrenargic neuron in several subpopulations throughout the pons and medulla [1, 2]. DREADD is a mutated G-Protein coupled receptor that is exclusively activated by an otherwise pharmacologically inert ligand, CNO[2]. This study sheds light on the functional projections that are associated with a subpopulation of noradrenergic cells that have neverbefore been functionally studiedMethod
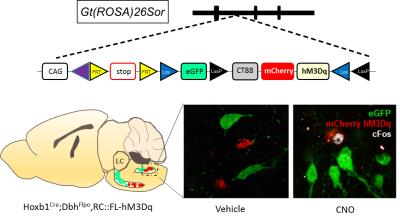
Male C57BL6;129 Hoxb1:cre; Dbh; FLPo; RC::FL-hM3Dq mice (n=8) genetically expressing hM3Dq and mCherry (Red) in r4 (Hoxb1cre) noradrenargic neurons were used (Figure 1). Modulation of these hM3Dq expressing neurons by CNO was confirmed using c-Fos immunohistochemistry (Figure 1). For PET experiments, mice were initially anesthetized under 1-3% isoflurane and injected with either saline or CNO (1 mg/kg, i.p.) in separate sessions. After 5 min, 18F-FDG was injected, followed by an uptake period where the mice were awake and allowed to resume their normal cage activities. After 45 minutes, the mice underwent a 10 min CT and 30 min PET scan in static mode. All PET data were analyzed using PMOD. For fMRI studies, mice were initially anesthetized using 1-3% isoflurane and thereafter, maintained under light anesthesia (0.75-1% isoflurane) while maintaining physiological homeostasis. Cerebral blood volume (CBV)-weighted fMRI responses were recorded by injecting a bolus dose of an in-house iron oxide nanoformulation (30 mg Fe/kg, i.v.) [3]. Single shot GE-EPI sequences (TR/TE = 3000/7.9 ms, matrix = 64x64, FOV = 1.92 cm2, slice number = 26 and slice thickness = 0.3 mm. were acquired using Bruker 9.4T MR scanner with a 72 mm quad-transmit only volume coil and a quad-receive only mouse brain coil. We acquired fMRI data continuously for 40 min, with CNO administered 10 min after scan onset. Isotropic 3D fMRI data were motion corrected, aligned to a baseline EPI population atlas using AFNI-based pipeline and manually skull stripped on ITK-SNAP. Independent Component Analysis (ICA) was performed on fMRI data using 20 IC maps with FSL MELODIC and rendered in 3D using AMIRA.Result and Discussions
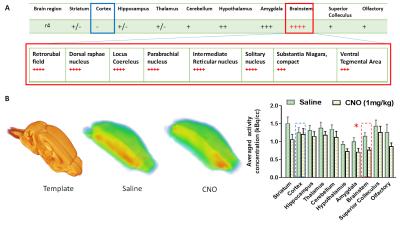
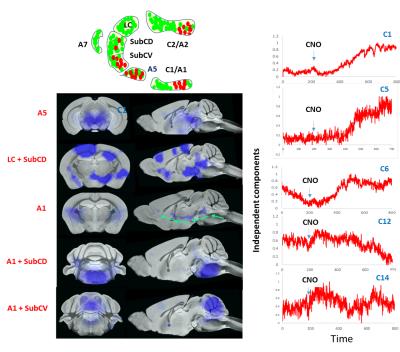
CNO administration augmented the firing of Hoxb1-derived noradrenargic neurons producing significant (P<0.05) decrease in glucose metabolism in the brainstem compared to basal glucose uptake on the same saline-treated subject (Fig 2). Consistent with PET data, ICA maps of Hoxb1-hM3Dq animals statistically thresholded at P<0.05 showed strong CNO-induced CBV decrease in brainstem, amygdala and hypothalamus (Fig 3). We, however, did not observe a similar trend in wild type controls. The first (C1), sixth (C6), twelfth (C12) and fourteenth (C14) components of the ICA maps corresponded to responses from A5, A1, A1 +Sub-CD and A5+Sub-CV anatomical nuclei respectively, all of which except Sub-CD, are densely populated with hM3Dq expressing HoxB1-noradrenargic neurons (Fig 3[SYI1] ) Among these, C6 revealed an interesting trend where the signal seemed to have originated from A1 and travelled all the way to cortex via medial forebrain bundle fibers that transmits information between VTA and nucleus accumbens. A careful analysis of all IC components also revealed little to moderate effects in various forebrain and midbrain areas including caudate putamen, which as per conventional histology based tract tracing, does not seem to share projections with Hoxb1-noradrenargic neurons. Despite the fact HoxB-noradrenargic neurons sparsely innervates with cortex and cerebellum, C5 and C14 revealed some effect of CNO in these regions. In particular, signal onset from C5 was relatively delayed suggesting that these regions may be activated via a secondary input from Locus Coereleus derived noradrenargic neurons having very strong and dense innervations with HoxB 1-derived noradrenargic neurons and moderate to strong projections into cerebellum and cortex respectively.Conclusion
We developed a novel chemogenetic based fMRI approach that opens a new avenue for mapping functional projections of genetically distinct neuronal subpopulation. For the first time, we showed that ICA-based fMRI analaysis is sensitive enough to dissect the complex polysynaptic pathways associated with chemogenetic modulation of Hoxb-1 derived noradrenargic neurons. This method will contribute a novel toolbox for dissecting the contribution of individual subpopulations of noradrenargic neurons to circuits regulating complex behavioral and physiological processes.Acknowledgements
We thank members of the Shih lab for valuable discussions concerning the experiments described in this manuscript. M.D was supported by CDF-HFSP. [SYI1] Y.Y.I.S. was supported by NINDS R01 NS091236, NIMH R01 MH111429, R41 MH113252, R21 MH106939, NIAAA U01 AA020023, R01 AA025582, American Heart Association 15SDG23260025, and is an Ellen Schapiro & Gerald Axelbaum Investigator and Young Investigator Award recipient of the Brain & Behavior Research Foundation. Dr. Patricia Jensen at NIEHS is duly acknowledged for providing Hoxb1-hm3Dq mice. Yu-Wei Chen is acknowledged for helping with immunohistochemical analysis.References
1. Robertson, S.D., et al., Developmental origins of central norepinephrine neuron diversity. Nature neuroscience, 2013. 16(8): p. 1016-1023.
2. Zhu, H. and B.L. Roth, DREADD: a chemogenetic GPCR signaling platform. International Journal of Neuropsychopharmacology, 2015. 18(1): p. pyu007.
3. Decot, H.K., et al., Coordination of Brain-Wide Activity Dynamics by Dopaminergic Neurons. Neuropsychopharmacology, 2016.
Figures