1039
Wave-CAIPI ViSTa: Accelerated mapping for direct visualization of myelin water1Center for Brain Imaging Science and Technology, Department of Biomedical Engineering, Zhejiang University, Hangzhou, People's Republic of China, 2Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Charlestown, MA, United States, 3Department of Radiology, Harvard Medical School, Boston, MA, United States, 4MR Collaboration NE Asia, Siemens Healthcare, Shanghai, People's Republic of China
Synopsis
Direct Visualization of Short Transverse relaxation time component (ViSTa) is a new myelin water imaging method, which directly extracts myelin water signal through double inversion RF pulses (DIR) to preserve signal from short T1 components. This method is robust and sensitive to demyelinated lesions, but suffers from extremely long scan time. Herein, we propose accelerated ViSTa acquisition through Simultaneous Multi-Slice (SMS) wave-CAIPI while keeping the high fidelity of ViSTa and MWF maps. As the average/maximum g-factor noise amplification is only 3/10% at MultiBand-4, further acceleration is likely to shorten whole brain MWF acquisition to within 5 minutes.
Introduction
MRI myelin water imaging has been used as a non-invasive tool for detecting integrity of white matter (WM), where T2 or T2* decay is measured and fitted to a multi-component WM model for myelin water percentage estimation (with the shortest T2 or T2*, e.g. T2<40 ms or T2*<25 ms at 3T) [1]. This method is often unstable due to low SNR of the heavily decayed signal and the ill-conditioned fitting procedure. Recently a method named direct Visualization of Short Transverse relaxation time component (ViSTa) was proposed [2], using double inversion RF pulses (DIR) to extract signal from short T1 components, which was suggested to be generated by myelin water. This method was successful in detecting MS lesions [2] while avoiding the instability from multi-component model fitting, indicating its potential for clinical use. However, the scan time for ViSTa was extremely long (about 3.1 min for a single-slice acquisition), hindering its wide application and deployment for clinical studies. Wave-CAIPI [3] is a recently proposed acceleration method that allows high reduction factor while achieving low g-factor noise penalty. In this study, we demonstrated the potential of accelerating ViSTa with Simultaneous Multi-Slice (SMS) wave-CAIPI, where the quality of ViSTa images were near identical after 4x acceleration. The low g-factor penalty from wave-CAIPI (gavg/gmax=1.03/1.10) warrants an even higher acceleration factor for rapid, whole brain ViSTa acquisition.Methods
The wave-CAIPI ViSTa sequence is shown in Fig 1. An SMS RF pulse with MultiBand rate (MB) of 4 with 24mm slice separation and 90 degree flip angle was applied for excitation. Scanning was performed on a 3T scanner (MAGNETOM Prisma, Siemens, Erlangen, Germany), with the following parameters: TI1 = 560ms, TI2 = 220ms and TD = 380ms; slice thickness 3mm, FOV 220×220mm2 with matrix size 192×192; TR = 1160ms, TE = 7.15ms, readout bandwidth 100Hz/px. Five datasets were acquired using sequence prototype:
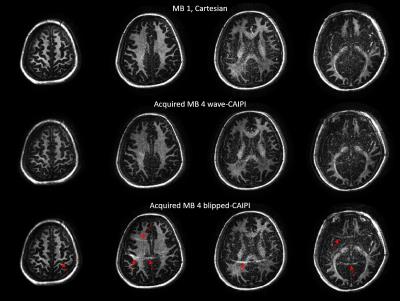
(1) MB=4 wave-CAIPI ViSTa, with 4 slices simultaneously acquired in 3 min 42 sec.
(2) MB=4 blipped-CAIPI ViSTa, with 4 slices simultaneously acquired (using shift of FOV/4 increment along PE direction between slices) in 3 min 42 sec.
(3) Fully sampled single slice ViSTa images on same location with MB = 1 excitation and conventional Cartesian readout as golden standard, with 4 slices acquired sequentially and each with acquisition time of 3 min 42 sec. These four slices were also collapsed with FOV/4 shift to form simulated MB=4 blipped-CAIPI ViSTa.
(4) Same MB = 1 ViSTa acquisition as (3) but with wave readout gradients, and then collapsed to form simulated MB=4 wave-CAIPI ViSTa.
(5) FLASH MB=1 acquisition, with the same parameter as (3) but with the DIR preparation turned off.
The images acquired with wave readout gradients (datasets 1 and 3) were reconstructed using the auto-calibrated wave-CAIPI method [4], while blipped-CAIPI images (datasets 2 and 4) were reconstructed with SENSE. These results were compared to the original fully sampled MB=1 results for the fidelity of the result of two kinds of CAIPI methods. Myelin water fraction (MWF) maps were calculated by dividing ViSTa by the FLASH data (dataset 5) and multiplying with an adjustment factor [2].
Results
Fig 2 demonstrates the results of simulated MB=4 for both with and without wave readout gradients, and their comparison with the corresponding original MB=1 images. The blipped-CAIPI images showed obvious residual aliasing artifacts mainly from scalp in neighboring slices, while no structured aliasing artifacts were observed in the images with wave readout. Note that the degradation of signal-to-noise ratio (SNR) in both cases should be attributed to sqrt(MB) increase in noise standard deviation in this simulation, which combines noisy data from 4 separate acquisitions to generate the collapsed slice. The g-factor maps in Fig. 2 proved that this is not due to the g-factor penalty alone, as gmax was only around 2 in SMS-CAIPI case. This sqrt(MB) amplification in noise would not be presented in the actual SMS acquisition.
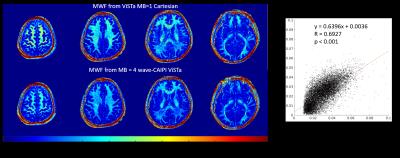
Fig 3 demonstrates a dramatic improvement of using wave-CAIPI SMS acquisition, showing almost no visible degradation when comparing with non-accelerated MB=1 case. Fig 4 shows the similarity of MWF maps calculated by the acquired MB=1 Cartesian and MB=4 wave-CAIPI data.
Discussion and Conclusion
ViSTa is a useful technique for direct observation of myelin contents with good reproducibility [5] but with extremely low acquisition efficiency. EPI readout could be one choice for acquisition [6], but the distortion in EPI and the effect of T2* decay through readout train (especially for short T2* of ViSTa signal) would inevitably introduce error to the subsequent MWF calculation. The high scalp signal is one of the challenges of using parallel imaging for ViSTa acceleration, leaving residual artifacts in blipped-CAIPI. Although it may be alleviated through fat saturation, the MT effect of saturation pulse also may affect the calculated MWF values [2].
In this study, we demonstrated the ability of wave-CAIPI in efficiently accelerating the acquisition while keeping the high fidelity of ViSTa and MWF maps. Further acceleration of ViSTa is likely to shorten the whole brain MWF acquisition to within 5 minutes.
Acknowledgements
The author (Zhe Wu) thanks Dr. Jongho Lee and Dr. Se-Hong Oh for their insightful discussion. This work was supported by National Natural Science Foundation of China (NSFC 81401473).References
[1] Dosik Hwang, Dong-Hyun Kim, Yiping P. Du. In vivo multi-slice mapping of myelin water content using T2 * decay. NeuroImage. 2010; 52: 198–204.
[2] Se-Hong Oh, Michel Bilello, Matthew Schindler, Clyde E. Markowitz, John A. Detre, Jongho Lee. Direct visualization of short transverse relaxation time component (ViSTa). NeuroImage 83 (2013) 485–492.
[3] Berkin Bilgic, Borjan A. Gagoski, Stephen F. Cauley, Audrey P. Fan, Jonathan R. Polimeni, P. Ellen Grant, Lawrence L. Wald, and Kawin Setsompop. Wave-CAIPI for Highly Accelerated 3D Imaging. Magnetic Resonance in Medicine 73:2152–2162 (2015).
[4] Stephen F. Cauley, Kawin Setsompop, Berkin Bilgic, Himanshu Bhat, Borjan Gagoski, and Lawrence L. Wald. Autocalibrated Wave-CAIPI Reconstruction; Joint Optimization of k-Space Trajectory and Parallel Imaging Reconstruction. Magnetic Resonance in Medicine 2016. Epub ahead.
[5] Han Jang, Yoonho Nam, Yangsoo Ryu, and Jongho Lee. Comparison of ViSTa myelin water imaging with DTI and MT. Proc. Intl. Soc. Mag. Reson. Med. 23 (2015): p7. [6] Se-Hong Oh, Sung Suk Oh, Joon Yul Choi, Jang-Yeon Park, and Jongho Lee. Three Dimensional Quantitative Myelin Water Imaging using Direct Visualization of Short Transverse Relaxation Time Component (ViSTa). Proc. Intl. Soc. Mag. Reson. Med. 22 (2014): p4280.
[6] Se-Hong Oh, Sung Suk Oh, Joon Yul Choi, Jang-Yeon Park, and Jongho Lee. Three Dimensional Quantitative Myelin Water Imaging using Direct Visualization of Short Transverse Relaxation Time Component (ViSTa). Proc. Intl. Soc. Mag. Reson. Med. 22 (2014): p4280.
Figures
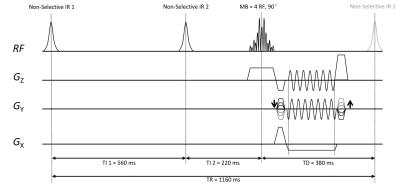
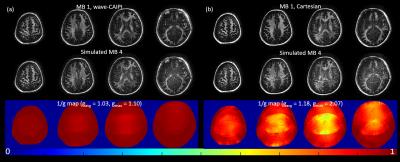
Figure 2 MB=1 images and the reconstructed slices from simulated MB=4 acceleration. Notice that the SNR degradation in the middle row should be attributed to the superimposition of noise from all slices according to g-factor map.
(a) Using data with wave readout gradients (dataset 4). Upper: Reconstructed MB=1 with wave encoding. Bottom: Reconstructed wave-CAIPI ViSTa images at MB=4 simulated by collapsing images in the upper row.
(b) Using normal Cartesian data (dataset 3). Upper: Original ViSTa images. Bottom: Reconstructed images from the simulated MB=4 SMS-CAIPI ViSTa. Note the obvious residual aliasing artifacts inside and outside cerebrum.