0887
Detection of in vivo biomarkers in fungal brain infection models and potential determination of cell viability.1Imaging and Pathology, University of Leuven, Leuven, Belgium, 2Laboratory of Clinical Bacteriology and Mycology, University of Leuven, Leuven, Belgium
Synopsis
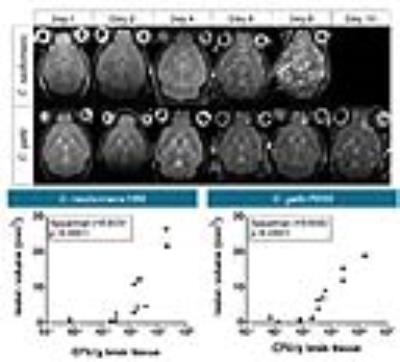
Animal models of cerebral infection by the pathogenic fungi Cryptococcus neoformans and C. gattii were developed and assessed longitudinally by using anatomical and diffusion MRI as well as MR spectroscopy. MR spectroscopy identified in vivo biomarkers for potential etiological diagnosis and more importantly for quantification of the fungal load in living animals. Our results have great potential to assist in the differential diagnosis of brain lesions in patients, whereby MR spectroscopy is a safer, non-invasive and rapid method in comparison to traditional invasive diagnostic methods such as CSF sampling or biopsies.
Introduction
Cryptococcus neoformans and C. gattii are encapsulated yeasts that can cause life-threatening fungal infections primarily affecting immunocompromised individuals (1). The main route of infection is by inhalation, but cryptococci can also spread from the lung to the brain. Thereby, they can cause lesions (cryptococcomas) that are difficult to distinguish from other pathologies (like cystic brain tumors or bacterial abscesses) by means of conventional radiological examinations such as computed tomography (CT) or magnetic resonance imaging (MRI). Although both cryptococcus species are related, potential differences in their clinical presentation and disease pathogenesis have been reported (2). Current techniques used in preclinical research, such as colony-forming unit (CFU) counting to determine fungal load are limited to snapshots of the disease and do not allow longitudinal studies of the same animal because of their invasive character. The aim of this study was to use MRI and MR spectroscopy (MRS) to identify in vivo parameters that can give a quantitative read-out of the fungal load in living animals at an anatomical and metabolic level over the course of disease progression. Furthermore, this approach was used to identify potential differences in pathogenesis between C. neoformans and C. gattii infections.Materials and Methods
Animal models
Two animal models were used, (1) Cryptococcus strains (C. neoformans H99 and C. gattii R265; 104 fungal cells) were stereotactically injected into the right striatum of female BALB/c mice to induce localized lesions and (2) Animals were injected intravenously with 5 x 104 cryptococcal cells to reflect disseminated disease. For local injections, mice were scanned at day 5, 8, 10 and 13 post infection. Mice that were infected by i.v. injection were scanned repeatedly for a period of 1 to 4 weeks.
Imaging
All imaging experiments were performed using a small animal 9.4 T MRI scanner (Biospec 94/20, Bruker Biospin, Ettlingern, Germany) equipped with 600mT/m gradients and using a 7cm linearly polarized resonator for transmission and an actively-decoupled mouse brain surface coil for receiving (Bruker Biospin). We acquired anatomical 3D MRI (RARE, TR/TE: 1000/36ms, 0.1mm resolution), 2D DWI (TR/TE: 3s/ 27ms, b-values: 30 to 1500ms,) and single voxel MRS (PRESS, 2 x 2 x 2mm3 voxel, TR/ TE: 1.8s/ 20ms). MR images were analyzed using Paravision 5.1. MRS results were quantified by using jMRUI to determine absolute metabolite concentrations (3).
Validation
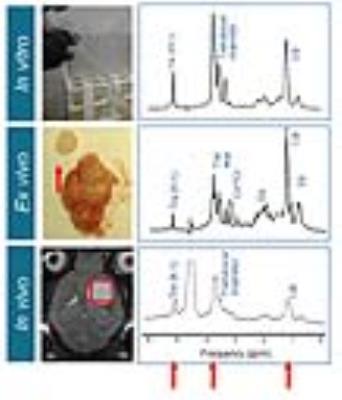
After the final time point, brains were removed for confirmation by histology, fungal load quantification by CFU counts and NMR spectroscopy. Brain metabolites were assigned and quantified using ex vivo NMR spectroscopy (400MHz Bruker) as described (4, 5).
Results
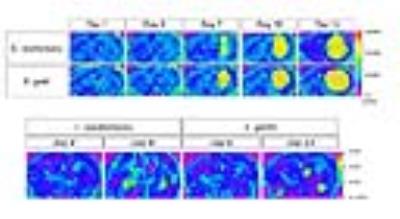
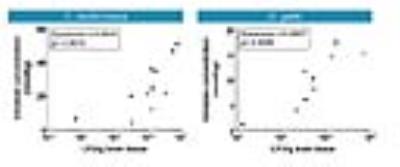
Disease progression was monitored longitudinally with MRI whereby increases in lesion size correlated with increased fungal load (Figure 1). Diffusion-weighted MRI showed increased diffusion inside the lesion compared to normal brain tissue (Figure 2). MRS detected the presence of typical fungal metabolites like trehalose and mannitol but also lipids inside the cryptococcal lesions, which was confirmed by ex vivo NMR spectroscopy (Figure 3). Trehalose concentrations correlated with the fungal load and a higher in vivo trehalose production per cell by C. gattii compared to C. neoformans was revealed (Figure 4). Based on MRS based detectability limits 104 and 105 cfu per gram tissue were detectable for C. gattii and C. neoformans, respectively. MRI was also applied to study infection in the disseminated disease model. Hereby, a significantly higher amount of brain lesions was detected for C. neoformans infected animals compared to C. gattii infected animals (Figure 5).Discussion
For the first time, this study presents a method for non-invasive follow-up and characterization of cerebral cryptococcomas in a mouse model. While anatomical MRI and DWI was able to monitor disease progression, MRS identified trehalose as a characteristic metabolite that can be used for diagnostic purposes. Comparison with MRS-based metabolite profiles from previously published models of cystic glioblastoma or bacterial abscesses allows non-invasive distinction between those pathologies from cryptococcoma and hereby identifies the lesion-causing pathogen (5, 6). In addition, trehalose may act as a quantitative biomarker for in vivo assessment of the viable fungal load. Currently, experiments are in progress to further assess this hypothesis on more cryptococcal strains and in treatment models. Our current findings have great potential to assist in the differential diagnosis of brain lesions in patients, whereby MRS is a safe, non-invasive and rapid method in comparison to traditional methods such as CSF sampling or biopsies (7).Acknowledgements
We are thankful for financial support from the European Commission for EU MC ITN TRANSACT 2012 (no. 316679), for the Infect-ERA project CryptoVIEW and from the Flemish Government FWO for the project G.0869.12N.
References
1. Del Poeta M, Casadevall A. Ten challenges on Cryptococcus and cryptococcosis. Mycopathologia. 2012, 173:303–10
2. Chen SCA, Meyer W, Sorrell TC. Cryptococcus gattii infections. Clin. Microbiol. Rev. 2014, 27:980-1024
3. Osorio-Garcia MI, Sima DM, Nielsen FU, Himmelreich U, Van Huffel S. Quantification of magnetic resonance spectroscopy signals with lineshape estimation. J. Chemometrics 2011; 25: 183–192
4. Croitor-Sava A, Beck V, Sandaite I, Van Huffel S, Dresselaers T, Claus F, Himmelreich U, Deprest J. High-resolution 1H NMR spectroscopy discriminates amniotic fluid of fetuses with congenital diaphragmatic hernia from healthy controls. J. Proteome Res. 2015, 14: 4502−4510
5. Himmelreich U, Dzendrowskyj T, Allen C, Dowd S, Malik R, Shehan P, Russell P, Mountford CE, Sorrell TC.: Cryptococcomas distinguished from gliomas with magnetic resonance spectroscopy : an experimental rat and cell culture study; Radiology 2001, 220: 122-128
6. Himmelreich U, Accursso R, Malik R, Dolenko B, Somorjai RL, Gupta RK, Gomes L, Mountford CE, Sorrell TC. Identification of Staphylococcus aureus brain abscesses: rat and human studies using H-1 MR Spectroscopy. Radiology 2005, 236: 261-270
7. Sorrell TC, Wright LC, Malik R, Himmelreich U. Application of proton nuclear magnetic resonance spectroscopy to the study of Cryptococcus and cryptococcosis. FEMS Yeast Research 2006, 6: 558-566
Figures