0874
Diffusion Spectrum Imaging Tractography of the Human Tongue1Diagnostic Radiology, University of Maryland School of Medicine, Baltimore, MD, United States, 2Neural and Pain Sciences and Orthodontics, University of Maryland Dental School, Baltimore, MD, United States, 3Electrical and Computer Engineering, Johns Hopkins University, Baltimore, MD, United States
Synopsis
The human tongue is known to have a complex architecture of muscles. To fully understand how the muscle fibers are connected and how their relative position could affect the tongue functionality, diffusion weighted imaging is needed. Diffusion Spectrum Imaging (DSI) is able to characterize the fiber structure on a sub-voxel level including the fiber crossing or branching. As DSI sequences are usually time-consuming, in-vivo studies using DSI can be challenging. In this study, we present for the first time DSI of the post-mortem human tongue. and the associated tractography delineating the tongue muscle fibers. DSI has the ability to identify fiber crossings within the human tongue.
Purpose
The human tongue is known to have a complex architecture of muscles [1]. To fully understand how the muscle fibers are connected and how their relative position could affect the tongue functionality, diffusion weighted imaging is needed. Diffusion tensor imaging (DTI) can trace some of the fiber tracts of the human tongue noninvasively. However, tongue fiber crossings or branching could not be resolved using DTI. Diffusion Spectrum Imaging (DSI) is able to characterize the fiber structure on a sub-voxel level including the fiber crossing or branching. As DSI sequences are usually relatively time-consuming, in-vivo studies using DSI can be challenging. In this study, we present DSI of the post-mortem human tongue, and the associated tractography delineating the tongue muscle fibers.Methods
Diffusion imaging in the tongue was performed on a post mortem 73-year-old female body, around 36 hours after death. The diffusion images were acquired on a Siemens TrioTim scanner using a Readout-Segmented Echo-Planar diffusion sequence (RESOLVE) [2], with TE=125 ms, and TR=3700 ms.A diffusion spectrum imaging scheme was used with a total of 202 diffusion directions at a maximum b-value of 4000 s/mm2.The in-plane resolution was 2.5 mm. The slice thickness was 2.5 mm. The diffusion data were reconstructed using diffusion spectrum imaging [2] reconstruction with a Hanning filter of 8. A deterministic fiber tracking algorithm [3] was used with an angular threshold of 42⁰, step size of 0.55 mm, and the anisotropy threshold of 0.035. The fiber trajectories were smoothed by averaging the propagation direction with 69% of the previous direction. Tracts with length less than 7 mm were discarded. The analysis was conducted using DSI Studio [4].
Results
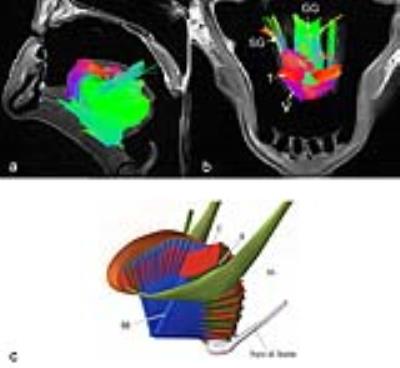
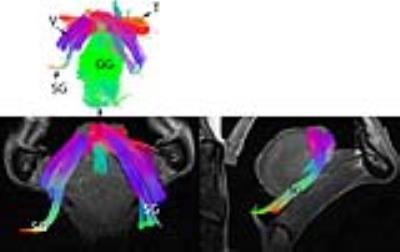
A few key tongue muscle fiber tracts are shown in Fig 1(a-b), as compared to the model shown in Fig. 1.c. Color coding for the tracts are the same for all figures; anterior-posterior is depicted by a green color. Superior-inferior is blue, and transverse direction is red. The tractography images presented in Fig. 1-2 show detailed information about the direction of different muscle fibers. We could identify the fiber tracts of genioglossus (GG), styloglossus (SG), vertical (V), and transverse (T) muscles.Discussion
This study shows for the first time the use of DSI to successfully delineate the complex muscle fibers of the human tongue. DSI has the ability to identify fiber crossings within the human tongue It is known that tissue properties change after death due to tissue decomposition, which leads to a decrease of the T2 relaxation time in the ex-vivo samples, as well as a change in the diffusion properties. These factors make the time of scan after death to be very critical. We show here that within a post-mortem time of 2 days we can acquire reasonable DSI data in the tongue. We will continue to optimize the post mortem DSI for better identification of human tongue muscle.Conclusion
The tongue has a complex muscle architecture that could be resolved using DSI. The fiber crossing and fiber branching of the tongue muscles are essential for understanding the different physiological functions of different parts of the tongue. This study shows the feasibility of precise delineation of different human tongue muscle fibers using DSI.Acknowledgements
No acknowledgement found.References
[1] M. Stone, J. Woo, J. Lee, T. Poole, A. Seagraves, M. Chung, E. Kim, E. Z. Murano, J. L. Prince and S. S. Blemker, "Structure and variability in human tongue muscle anatomy," Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization, pp. 2168-1171, 2016.
[2] D. A. Porter and R. M. Heidemann, "High Resolution Diffusion-Weighted Imaging Using Readout-Segmented Echo-Planar Imaging, Parallel Imaging and a Two-Dimensional Navigator-Based Reacquisition," Magn Reson Med, vol. 62, pp. 468-475, 2009.
[3] V. J. Wedeen, P. Hagmann, W. Y. Tseng, T. G. Reese and R. M. Wisskoff, "Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging.," Magn Reson Med, vol. 54, no. 6, pp. 1377-86, 2005.
[4] F.-C. Yeh, T. D. Verstynen, Y. Wang, J. C. Fernández-Miranda and W.-Y. I. Tseng, "Deterministic diffusion fiber tracking improved by quantitative anisotropy," PLoS ONE, vol. 8, no. 11, p. e80713, 2013.
[5] "http://dsi-studio.labsolver.org," [Online]. [6] H. Takemoto, "Morphological analyses of the human tongue musculature for three-dimensional modeling.," J Speech Lang Hear Res, vol. 44, no. 1, pp. 95-107, 2001.
Figures