0836
Quantifying neuronal microstructure integrity with TE dependent Diffusion Imaging (TEdDI)1Center for Biomedical Imaging, New York University School of Medicine, New York, NY, United States
Synopsis
We resolve degeneracies in the estimation of parameters of white matter integrity by exploiting the observed echo time dependency of diffusion MRI signal in human brain white matter. Empirical results and statistical analyses reveal that adding compartment-specific T2 relaxation times to biophysical models of diffusion MRI improves the parameter estimation in terms of precision and accuracy.
Purpose
To resolve degeneracies in diffusion MRI parameter estimation of neuronal microstructure, and to determine compartmental $$$T_2$$$ relaxation times.
Biophysical modeling of the diffusion-sensitized MRI (dMRI) signal is the way to achieve specificity, not just sensitivity, to tissue microstructure. Unfortunately, parameter estimation for dMRI models is degenerate.1,2 In particular, it has been shown empirically and analytically that (at least) two branches of model parameters, often both biophysically plausible, fit dMRI data equally well. Here we report a nontrivial dMRI signal dependence on echo time (TE) in human brain in vivo. We demonstrate, empirically and theoretically, that this TE dependence, which we interpret as originating from compartment-specific $$$T_2$$$ values3,4, is a promising "orthogonal measure" able to break the degeneracy in parameter estimation, and also yield important relaxational metrics.
Methods
We extended a widely used two-compartment biophysical model2,5-7 of water inside narrow impermeable "sticks" representing axons, embedded in an extra-cellular matrix, by including the compartment-specific $$$T_2$$$-relaxation parameterized by $$$T_2^a$$$ and $$$T_2^e$$$ for the axonal and extra-axonal compartments, respectively: $$S_\mathbf{\hat{g}}\mathrm{(}b,\,T_E)\,=\,S(0,0)\cdot\int_{|\mathbf{\hat{n}}\mathrm{|}=1}\,d\mathbf{\hat{n}}\mathcal{P}\mathrm{(}\mathbf{\hat{n}}\mathrm{)}\mathcal{K}\mathrm{(}b,T_E,\mathbf{\hat{g}}\cdot\mathbf{\hat{n}}\mathrm{)},\quad(1)$$
with
$$\mathcal{K}(b, T_E, \xi)=fe^{-T_E/T_2^a-bD_a\xi^2}+(1-f)e^{-T_E/T_2^e-bD_e^\perp\,-b(D_e^\parallel-D_e^\perp)\xi^2}\quad(2)$$
being the signal response of a individual fiber fascicle, and $$$S(0,0)$$$ the proton density. We adopt the recently introduced rotationally invariant framework2,7 that allows limiting the dimensionality of the parameter space by factoring out the axonal orientation distribution function (ODF) $$$\mathcal{P}({\bf\hat{n}})$$$. Basically, we solve an equivalent system
$$S_{l}(b,T_E;x)=p_l\,K_l(b,T_E;x)\quad(3)$$
where rotational invariants of the signal and ODF
$$S_l^2=\sum_{m=-l}^l |S_{lm}|^2/4\pi(2l+1),\quad\mathrm{and}\quad\,p_l^2=\sum_{m=-l}^l |p_{lm}|^2/4\pi(2l+1),\quad(4)$$
one for each $$$l$$$, are constructed from their spherical harmonics coefficients. The projections $$$K_l(b,T_E;x)$$$ of kernel (2) onto Legendre polynomials include explicit $$$T_E$$$ dependence. Here $$$x$$$ denote scalar model parameters including $$$T_2^a,\,T_2^e$$$, axonal volume fraction ($$$f$$$), intra-axonal diffusivity ($$$D_a$$$), parallel and perpendicular extra-axonal diffusivity ($$$D_e^\parallel$$$ and $$$D_e^\perp$$$). The spherical mean technique8,9 effectively corresponds to a constrained version of the $$$l=0$$$ rotational invariant, $$$K_0$$$. Here we include up to the 2nd order invariant2,7 $$$K_{0,2}$$$, involving the axonal dispersion $$$p_2$$$, which provides an important extra equation, and do not apply any constraints on scalar parameters $$$x.$$$ Including $$$K_{0,2}$$$, together with experimentally varying $$$T_E$$$, constitutes a novel parameter estimation framework which we refer to as TEdDI, TE dependent Diffusion Imaging.
Data
A healthy volunteer underwent imaging on a Siemens Prisma 3T MR scanner using a 64-channel receiver head coil with an EPI readout diffusion-weighted sequence. Monopolar diffusion gradients were applied along 30 isotropically distributed directions for $$$b=0.5,\,1,\,2,\,3,\mathrm{and}\,4\,ms/\mu\,m^2$$$. This diffusion protocol was repeated for $$$T_E\,=\,69,\,90,\,120,\,\mathrm{and}\,150\,\mathrm{ms}$$$. Additionally, 54 non-diffusion weighted images were acquired with $$$T_E$$$’s equidistantly distributed between 55 and 145$$$\,ms$$$. The following imaging parameters were kept constant: $$$\mathrm{TR}: 4000\,\mathrm{ms}$$$, in-plane resolution: $$$2.5\times\,2.5\,\mathrm{mm^2}$$$, slice thickness: $$$2.5\,\mathrm{mm}$$$.Results
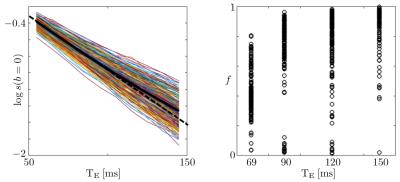
Figure 1: The logarithm of the non-diffusionweighted signal, and the estimated $$$T_2$$$-weighted axonal fraction are shown as function of $$$T_E$$$ for all voxels of the posterior limb of the internal capsule (PLIC). The non-monoexponential signal decay and monotonically changing fraction support the hypothesis of having compartment-specific $$$T_2$$$'s in the white matter (WM). Other ROIs, including corpus callosum (CC), show similar trends (unlike10).
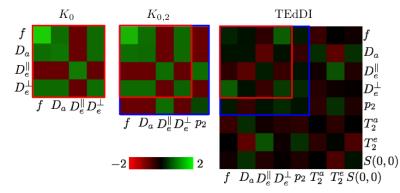
Figure 2: A statistical analysis of TEdDI using the inverse of the Fisher matrix, the covariance matrix of the model variables, predicts that by exploiting the $$$T_E$$$ dependence of the dMRI signal, the coefficient of variation of the model parameters lowers and, at least equally important, the correlation between parameters (e.g. $$$f$$$ and $$$D_a$$$) decreases. The analysis depends on a set of reference values for the model parameters, signal-to-noise ratio (SNR), and acquisition protocol. We adopted these settings from our clinical data.
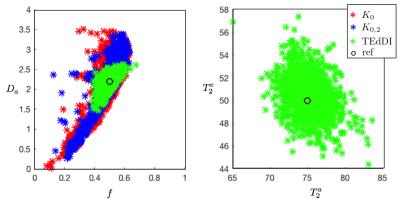
Figure 3: The decoupling of parameters, e.g. $$$f$$$ and $$$D_a$$$, turns out to be essential to overcome the degeneracy of the biophysical models. Simulations with random noise realizations and optimization initializations confirm these statistical results. Our analysis reveals that compartment $$$T_2$$$’s vary widely across different WM structures of interest.
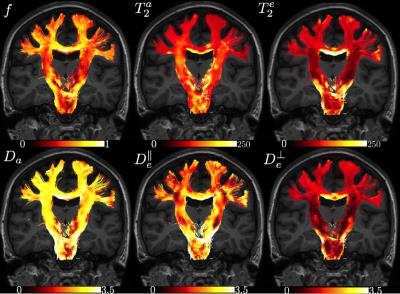
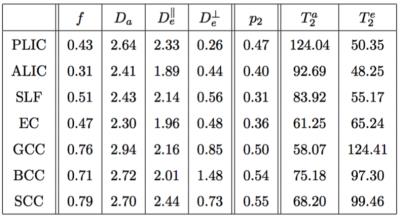
Maps are shown in Figure 4 and average ROI values are tabulated in Figure 5. Partial voluming with cerebrospinal fluid (CSF) that is characterized with high $$$T_2$$$ render the two-compartment model inaccurate around e.g. the CC/CSF border.
Discussion & Conclusion
$$$\bullet$$$ Empirical evidence is given for the $$$T_2$$$ values of intra- and extra-axonal water to be different.
$$$\bullet$$$ While more detailed study is needed, the tract-specific compartmental $$$T_2$$$’s may become valuable parameters in studying WM microstructure.
$$$\bullet$$$ The observed heterogeneity in $$$T_2$$$ values is not yet well understood, but already highlights the potential biases that might arise from assuming prior knowledge or globally fixing parameters, e.g. for kernel-driven diffusion methods such as constrained spherical deconvolution.
$$$\bullet$$$ Exploiting the $$$T_E$$$ dependence by including the compartmental $$$T_2$$$’s in the biophysical models bypasses several intrinsic problems of parameter estimation, resolving the previously highlighted degeneracies1,2.
Acknowledgements
Research was supported by the Fellowship from Raymond and Beverly Sackler Laboratories for Convergence of Physical, Engineering and Biomedical Sciences, by the Litwin Foundation for Alzheimer’s Research, and by the National Institute of Neurological Disorders and Stroke of the NIH under award number R01NS088040. JV is a Postdoctoral Fellow of the Research Foundation - Flanders (FWO; grant number 12S1615N).References
1. Jelescu et al., NMR Biomed 29, 33 (2016)
2. Novikov et al., Proc. ISMRM 2015, p.469; preprint https://arxiv.org/abs/1609.09144
3. Harkins et al. MRM 76(3): 793-800 (2012)
4. De Santis et al. ISMRM abstract 1998 (2016)
5. Jespersen et al., NeuroImage 34, 1473 (2007)
6. Jespersen et al., NeuroImage 49, 205 (2010)
7. Reisert et al., NeuroImage, DOI:10.1016/j.neuroimage.2016.09.058
8. Kaden et al., MRM 75, 1752-1763 (2016)
9. Kaden et al., NeuroImage 139, 346-359 (2016)
10. Ferizi et al. NeuroImage 118: 468-483 (2015)
Figures