0771
cBEaST: Cerebellar Brain Extraction based on Nonlocal Segmentation Technique – A comparison with state-of-the-art methods1Medical Physics Group, Institute for Diagnostic and Interventional Radiology, Jena University Hospital – Friedrich Schiller-University, Jena, Germany, 2Section of Experimental Neurology, Department of Neurology, Essen University Hospital, Germany, 3Erwin L. Hahn Institute for Magnetic Resonance Imaging, University Duisburg-Essen, Essen, Germany
Synopsis
An automatic segmentation of the cerebellum is required to determine the cerebellar volume and for improving spatial normalization in voxel-based analysis approaches. While existing segmentation approaches typically work quite robust in healthy subjects, errors in segmentation increase with cerebellar atrophy and typically require manual corrections. We introduce a novel cerebellum segmentation approach, referred to as cBEaST, that relies on a dedicated multi-resolution segmentation library with manually edited cerebellar masks of both healthy and diseased subjects in combination with multi-atlas-propagation and segmentation as implemented in BEaST. Finally segmentation of the cerebellum with BEaST is compared with the alternative techniques SUIT and FreeSurfer.
Introduction
The cerebellum is involved in a multitude of
disorders including multiple sclerosis1,2, schizophrenia3,4 and ataxia5,6. To characterize cerebellar involvement the degree
of cerebellar atrophy or alterations in functional, structural or quantitative
MR images via voxel-based analyses may be determined.5 Both of these strategies rely on an accurate
segmentation of cerebellar tissue for determining the cerebellar volume and for
improving spatial normalization7. Several approaches for cerebellum segmentation
exist including SUIT7 or Freesurfer8. While these approaches typically work quite robust
in healthy subjects, errors in segmentation increase with cerebellar atrophy
and typically require manual corrections. In this contribution, we set up a
segmentation library with manually edited cerebellar masks of healthy subjects
and patients with severe cerebellar atrophy to perform cerebellum segmentation
using multi-atlas-propagation and segmentation (MAPS) as implemented in the
BEaST (brain extraction based on nonlocal segmentation technique) toolbox.9 The novel approach is referred to as cBEaST and its
segmentation results are compared with the ones obtained with SUIT and
FreeSurfer.
Materials and Methods
Data Acquisition: Ninety-three participants (18 healthy subjects, 75 patients with different types of non-hereditary and hereditary degenerative cerebellar ataxia) underwent T1-weighted imaging with an MP-RAGE sequence (TI/TE/TR/BW=1100ms/3.26ms/2530ms/200Hz/px, acquired voxel size = 1mm×1mm×1mm) on a 3T-MRI system.
Established Cerebellar Segmentation: Cerebellar segmentation was performed using the SUIT toolbox (version 3.1: suit_isolate, version 3.2: suit_isolate_seg) and Freesurfer (recon-all). In the Freesurfer approach, the labeled partitions of gray and white matter of the left and right hemispheres as well as the brain stem were merged to reveal the segmentation of the cerebellum. Since among the three methods Freesurfer yielded segmentation results most closely to the cerebellar morphology, these ones were if necessary manually corrected to serve as reference segmentation for each subject.
cBEaST: Based on the data of all participants a library for non-local segmentation was set up containing T1w images and corresponding manually edited cerebellum segmentations at multiple resolutions (1mm, 2mm, and 4mm). To this end, the individual MR images and segmentation results were linearly registered to the SUIT template7 and down-sampled accordingly. Prior to down-sampling, bias field correction and image intensity normalization were applied to the T1-weighted images to justify similar alignment and signal intensity distributions among all datasets. Utilizing this specific library BEaST was then applied with the parameter configuration (resolution steps 4 mm, 2 mm and 1 mm) proposed in.9
Evaluation: All four segmentation methods were evaluated with respect to the manually edited reference segmentation by false positive projection mapping9 and the DICE coefficient10. To this end, the inferior and superior parts of the brainstem were excluded manually due to its different definitions in the investigated methods.
Results
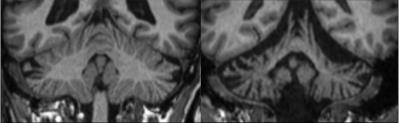
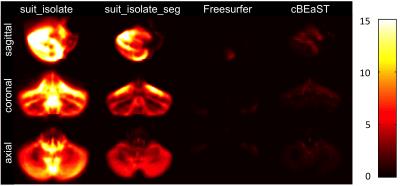
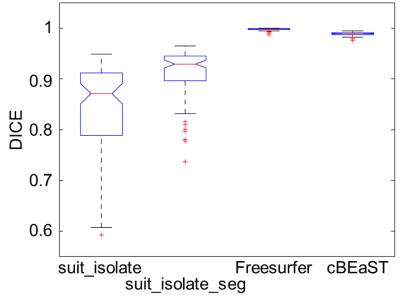
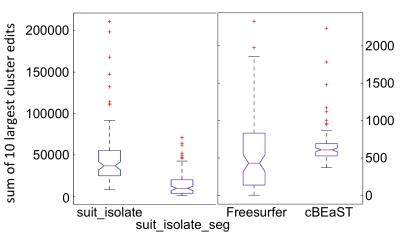
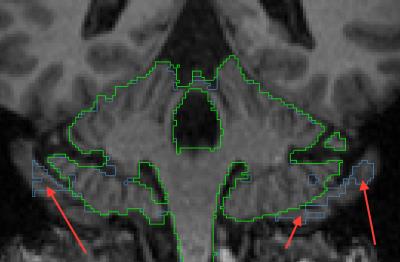
Exemplary, Figure 1 illustrates T1-weighted images of a healthy subject with a normal cerebellum and a patient with severe cerebellar atrophy. Figure 2 shows the maximum intensity projection of the false positives for the four different segmentation approaches. Both SUIT techniques revealed segmentations that extended to non-cerebellar structures (over-estimation), whereas both cBEaST and Freesurfer resulted in more accurately segmented cerebelli. The lowest deviations to the manually edited reference segmentations provided by the Freesurfer approach is putatively due to the fact that the Freesurfer results served as input to generate the reference segmentation. A similar finding on segmentation accuracy is provided by analyzing the Dice coefficient for the different segmentations that revealed highest median Dice coefficients with smallest distributions for the Freesurfer and cBEaST approaches. To assess the extending of the segmentation in non-cerebellar structures in more detail we plotted distributions of the sum of the 10 largest concatenated false positive clusters of each dataset and segmentation method (Fig. 4). While Freesurfer exhibits the best median performance the variance of the segmentation results is much larger compared to that of cBeast. Manual inspection revealed that only 5 datasets computed with cBEaST exhibited substantial deviations from the manual segmentation, while Freesurfer resulted in more than 15 subjects with large false positive clusters.Discussion
We demonstrated that current available methods for cerebellum segmentation are not suitable to be applied in patient populations with severe cerebellar atrophy without manual intervention. Although Freesurfer provides nearly an optimal segmentation, it fails on a significant amount of subjects (15 of 93) by over-estimating especially the inferior part (Fig. 5). To overcome these issues, we propose the use of a dedicated template library, which contains normal and severely atrophied cerebelli, in a multi-resolution non-local segmentation framework (cBEaST). Further extension of the library by additional subjects or MRI contrasts and optimization of the BEaST parameters is expected to further improve segmentation accuracy of cBEaST to finally overcome manual interventions.Acknowledgements
This work was supported by the German Research Foundation (DFG, DE2516/1-1, TI239/17-1).References
1. Weier K, Banwell B, Cerasa A, et al.
The role of the cerebellum in multiple sclerosis. Cerebellum. Jun 2015;14(3):364-374.
2. Sarica A, Cerasa A, Quattrone A. The
neurocognitive profile of the cerebellum in multiple sclerosis. Int J Mol Sci. May 28 2015;16(6):12185-12198.
3. Andreasen NC, Pierson R. The role of
the cerebellum in schizophrenia. Biological
psychiatry. Jul 15 2008;64(2):81-88.
4. Yeganeh-Doost P, Gruber O, Falkai P,
Schmitt A. The role of the cerebellum in schizophrenia: from cognition to
molecular pathways. Clinics (Sao Paulo). 2011;66
Suppl 1:71-77.
5. Stefanescu MR, Dohnalek M, Maderwald S,
et al. Structural and functional MRI abnormalities of cerebellar cortex and
nuclei in SCA3, SCA6 and Friedreich's ataxia. Brain. May 2015;138(Pt 5):1182-1197.
6. Deistung A, Stefanescu MR, Ernst TM, et
al. Structural and Functional Magnetic Resonance Imaging of the Cerebellum:
Considerations for Assessing Cerebellar Ataxias. Cerebellum. Feb 2016;15(1):21-25.
7. Diedrichsen J. A spatially unbiased atlas
template of the human cerebellum. NeuroImage.
Oct 15 2006;33(1):127-138.
8. Desikan RS, Segonne F, Fischl B, et al.
An automated labeling system for subdividing the human cerebral cortex on MRI
scans into gyral based regions of interest. NeuroImage.
Jul 1 2006;31(3):968-980.
9. Eskildsen SF, Coupe P, Fonov V, et al.
BEaST: brain extraction based on nonlocal segmentation technique. NeuroImage. Feb 1 2012;59(3):2362-2373.
10. Dice LR. Measures of the Amount of
Ecologic Association between Species. Ecology.
1945;26(3):297-302.
Figures