0702
In vivo 7 Tesla probabilistic neuroimaging structural atlas of human pedunculotegmental, oral pontine reticular and paramedian-raphe nuclei1Department of Radiology, A.A. Martinos Center for Biomedical Imaging, MGH and Harvard Medical School, Boston, MA, United States, 2Department of Neurosurgery, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, United States, 3Medical Physics Section, Department of Biomedicine and Prevention, Faculty of Medicine, University of Rome “Tor Vergata”, Rome, Italy, 4Department of Neurology, A. A. Martinos Center for Biomedical Imaging, MGH and Harvard Medical School, Boston, MA, United States, 5Department of Anesthesia, Critical Care and Pain Medicine, MGH, Boston, MA, United States
Synopsis
Mesopontine-tegmental nuclei such as the pedunculotegmental, oral-pontine-reticular and paramedian-raphe nuclei modulate arousal and motor functions. Dysfunction of these nuclei is implicated in the pathogenesis of disorders of consciousness, sleep disorders, and neurodegenerative diseases. However, a stereotaxic probabilistic atlas of these nuclei in humans does not exist. We used segmentation of 1.1 mm-isotropic 7 Tesla diffusion-fractional-anisotropy and T2-weighted images to generate and validate an in vivo probabilistic neuroimaging structural atlas of these nuclei in MNI space. We constructed this atlas to aid the localization of these nuclei in conventional images in future research and clinical studies of arousal and motor functions.
Introduction
Mesopontine tegmental nuclei such as the pedunculotegmental (PTg, also known as pedunculopontine), oral pontine reticular (PnO, also known as pontis oralis) and paramedian-raphe (PMnR) nuclei, are critical for arousal (e.g. wakefulness and REM sleep) and motor functions (e.g. locomotion) [1-4]. They are involved in the pathogenesis of disorders of consciousness [1], as well as sleep disorders [2] and neurodegenerative diseases [3-4]. For example, the PTg is a promising new target of deep brain stimulation in Parkinson's disease [3]. Nevertheless, a stereotaxic probabilistic atlas of these nuclei in living humans does not exist.
Purpose
To create a stereotaxic neuroimaging structural atlas of the left and right PTg (PTgl, PTgr), left and right PnO (PnOl, PnOr), and PMnR by the use of: 1) cutting-edge technology (7 Tesla scanner, 32-channel receive coil-array), which enabled us to push the current limits of MRI sensitivity; 2) a high-resolution (1.1 mm isotropic) multi-contrast (diffusion fractional anisotropy (FA) and T2-weighted) EPI approach, which provided exquisite complementary contrasts for brainstem anatomy with precisely matched geometric distortions and resolution.
Methods
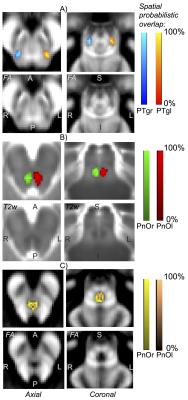
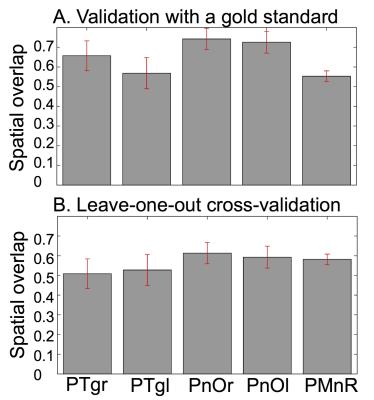
Data acquisition: Twelve subjects (6m/6f, age 28 ± 1) underwent 7 Tesla MRI under IRB approval. We adopted a common single-shot 2D EPI readout for 1.1 mm isotropic diffusion-tensor (DTI), and T2 weighted (T2w) sagittal images, with matrix size/GRAPPA factor/nominal echo-spacing = 180 × 240/3/0.82 ms. This yielded multi-contrast anatomical images with exactly matched resolution and geometric distortions. Additional MRI parameters were: spin-echo EPI, 61 slices, TE/TR = 60.8 ms/5.6 s, partial Fourier: 6/8, unipolar diffusion-weighting gradients for DTI, 60 diffusion directions (b-value ~ 1000 s/mm2), 7 interspersed “b0” images (non-diffusion weighted, b-value ~ 0 s/mm2, which were also used as T2w MRI), 4 repetitions, acquisition time/repetition 6’43”. Data analysis: On a single-subject basis, M.B. performed semi-automatic segmentation of multi-contrast (FA maps, computed from DTI, and T2w) images by k-means clustering, using the procedure described in [5]. This yielded single-subject labels of PTgl/r, PnOl/r, PMnR. These single-subject labels were coregistered (as in [5]) to MNI space through high dimensional non-linear transformations (ANTs [6]), and a probabilistic atlas for these structures in MNI space was created using the transformed labels (highest probability = 100 % overlap across subjects). Atlas validation: An expert neuroanatomist (C.S.) manually segmented the same regions in each subject, yielding gold standard single-subject labels of these nuclei. The probabilistic nuclei atlas was validated by computing for each nucleus the spatial overlap (i.e. the volume of the intersection divided by the volume of the reference label) between: (i) single-subject labels (derived from the semi-automatic segmentation) and gold-standard labels (derived from the manual segmentation); (ii) each single-subject label and the probabilistic atlas label (thresholded at 35%) generated by averaging the labels across the other 11 subjects (leave-one-out cross validation). For each nucleus, the spatial overlap of (i) and (ii) was then averaged across subjects and displayed.Results
The probabilistic neuroimaging structural labels in MNI space of PTgl/r, PnOl/r, PMnR are shown in Figure 1. The spatial overlap computed to validate the atlas labels with gold-standard labels, as well as using the leave-one-out cross validation approach is displayed in Figure 2.Discussion
Our findings demonstrated the feasibility of delineating tiny mesopontine tegmental nuclei of the arousal and motor systems by semi-automatic segmentation of single-subject high-contrast and high-sensitivity MRIs at 7 Tesla. This extends a few previous reports [1,3] of single-subject manual localization of these nuclei in neuroimages based on the identification of anatomical landmarks. Crucially, our work also demonstrated the feasibility of generating a validated in vivo stereotaxic probabilistic neuroimaging atlas of these structures after precise coregistration to MNI space. This atlas complements existing in vivo neuroimaging atlases of other brain structures [7-9].Conclusion
We foresee the use of the generated probabilitic atlas of the PTg, PnO and PMnR to aid the localization of these nuclei in conventional (e.g. 3 T) images in future research studies of arousal and motor functions. Further, this atlas, upon coregistration to clinical MRI, might improve the accuracy of interventions (e.g. placement of deep brain stimulation electrodes), the evaluation of lesions and the assessment of connectivity pathways uderlying arousal and motor mechanisms in a broad set of disease populations (e.g. disorders of consciousness, sleep disorders and neurodegenerative diseases).Acknowledgements
NIH NIBIB K01EB019474; NIH NIBIB P41-RR014075.References
1. Edlow BL, Takahashi E, Wu O, et al.
Neuroanatomic connectivity of the human ascending arousal system critical to
consciousness and its disorders. J Neuropathol Exp Neurol. 2012;71:531-546.
2. Boeve BF, Silber MH, Saper CB, et al.
Pathophysiology of REM sleep behavr disorder and relevance to
neurodegenerative disease. Brain. 2007;130:2770-2788.
3. Zrinzo L, Zrinzo LV, Tisch S, et al.
Stereotactic localization of the human peduncolopontine nucleus: atlas-based
coordinates and validation of a magnetic resonance imaging protocol for direct
localization. Brain. 2008;13:1588-1598.
4. Braak H, Del Tredici K, Rub U, et al.
Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol
Aging. 2003;24:197-211.
5. Bianciardi M, Toschi N, Edlow BE, et al.
Toward an in vivo neuroimaging template of human brainstem nuclei of the
ascending arousal, autonomic, and motor systems. Brain Connect. 2015;5:597-607.
6. Avants BB, Tustison NJ, Song G, et al. A
reproducible evaluation of ANTs similarity metric performance in brain image
registration. Neuroimage. 2011;54:2033-2044.
7. Destrieux C, Fischl B, Dale A, et al.
Automatic parcellation of human cortical gyri and sulci using standard
anatomical nomenclature. Neuroimage. 2010;53:1-15.
8. Desikan RS, Segonne F, Fischl B, et al.
An automated labeling system for subdividing the human cerebral cortex on MRI
scans into gyral based regions of interest. Neuroimage. 2006;31:968-980.
9. Tzourio-Mazoyer N, Landeau B,
Papathanassiou D, et al. Automated anatomical labeling of activations in SPM
using a macroscopic anatomical parcellation of the MNI MRI single-subject
brain. Neuroimage. 2002;15:273-289.
Figures