0696
Laminar microstructure and subfield connectivity of the human hippocampus revealed with ultra-high field diffusion MRI at 11.7T1Department of Radiology, Johns Hopkins University, Baltimore, MD, United States, 2Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, United States, 3Department of Pathology, Johns Hopkins University, Baltimore, MD, United States
Synopsis
In this work, we demonstrate high-field (11.7T) 3D diffusion MRI (dMRI) of the fixed human hippocampal formation to investigate its internal organization. The results of this study show delineation of fine hippocampal microstructure and inner connectivity of the human hippocampal subfields based on HARDI. Further, we demonstrate 3D reconstruction of intra-hippocampal subfield connectivity, e.g. the trisynaptic hippocampal circuit, using fODF-based probabilistic tractography with combined high spatial and angular resolution, which has important future applications for mapping of pathology-induced changes in the hippocampus in Alzheimer’s disease and related neurological disorders.
Purpose
The hippocampal formation is a structure of acute interest in neuroimaging and is implicated in a wide array of neurological disorders including epilepsy and Alzheimer’s disease1. Understanding and detailed three-dimensional (3D) delineation of the complex hippocampal neuroanatomy is vital to studies of neuro-psychiatric disorders, but remains difficult. Typical in vivo MR resolutions in the human brain are too coarse to differentiate layers of the hippocampus. Conventional relaxation-based ex vivo MRI offers excellent gray and white matter contrast with high resolution2-4, but provides limited ability to delineate fine inner hippocampal connectivity. Consequently, 3D mapping of intra-hippocampal microstructure and subfield connectivity has remained technically challenging. In this work, we demonstrate ultra-high field diffusion MRI (dMRI) to investigate the internal microstructural organization of the fixed human hippocampus. Using combined high angular and spatial resolution, we show, for the first time, intricate intra-hippocampal neuronal circuitry and microstructure delineated with dMRI, revealing the 3D internal organization of the hippocampus in unprecedented detail.Methods
Whole intact hippocampus specimens from adult human cadaver brains (n=3) were immersion-fixed with 10% formalin. MR imaging was performed on a high-field 11.7T scanner (Bruker Biospin, maximum gradient strength=740 mT/m) using an 8-channel volume-array coil. High angular resolution diffusion imaging (HARDI) data were acquired using a 3D 12-shot segmented diffusion-weighted (DW)-EPI sequence with the following imaging parameters: TE/TR = 27/550 ms, 1 average, bandwidth = 210 kHz, diffusion gradient duration (δ)/separation (Δ) = 5/15 ms, and partial Fourier factor of 1.4. 40 diffusion-encoding directions with b-value = 4000 s/mm2 and 2 non-diffusion-weighted (b0) images were acquired at an isotropic resolution of 160 x 160 x 160 µm3 and scan time of 10 h. Parametric fractional anisotropy (FA) and direction-encoded color (DEC) maps were calculated from tensor fitting in IDL. Fiber orientation distribution functions (fODFs) were estimated from the HARDI data using constrained spherical deconvolution (CSD) in MRtrix5 with a maximum harmonic order (lmax) of 6. Track density images (TDI)6 were generated using fODF-based probabilistic tractography7 with a grid size of 40 µm.Results and Discussion
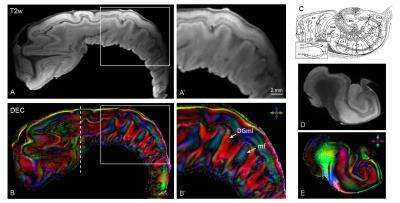
Mean SNR measurements in gray and white matter in the b0 images were 152±28 and 68±17, respectively. The dMRI results revealed intricate details of intra-hippocampal microstructure and circuitry (Fig. 1). DEC maps (Fig. 1B-B’) provided striking contrast to delineate the interleaved organization of mossy fiber axons (mf, blue) and apical dendrites of granule cells in the dentate gyrus molecular layer (DGml, red), based on their distinct orientations. In comparison, these features could not be effectively distinguished in non-diffusion-weighted or relaxation-based tissue contrasts (Fig. 1A-A’). Coronal slices in Fig. 1D-E further show the intricate anisotropic diffusion contrast driven by the intra-hippocampal neurite orientations (shown in Fig. 1C).
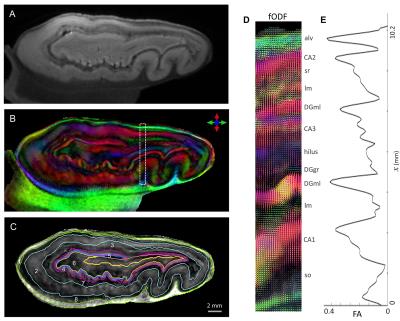
HARDI results enabled delineation of several laminar divisions and specific subfields of the hippocampus (Fig. 2). fODFs reconstructed using CSD (Fig. 2D) revealed a distinct laminar distribution. Fig. 2D shows fODFs across hippocampal gray matter reflecting the unique organization of axons and dendritic arbors in different layers, e.g., apical dendrites of granule and pyramidal cells in the DGml and cornus ammonis (CA1-CA3), and multiple peak components in the stratum lacunosum-moleculare (lm) in perfect agreement with known crossing fiber orientations of pyramidal cell apical dendrites and perforant path axons that innervate this layer. Quantitative FA plot in Fig. 2E exhibits a distinct laminar profile corresponding to microstructural heterogeneity across different layers. The intricate contrast provided by HARDI enabled detailed 3D segmentation of several hippocampal layers (Fig. 2C), which were not discernible in non-diffusion-weighted images.
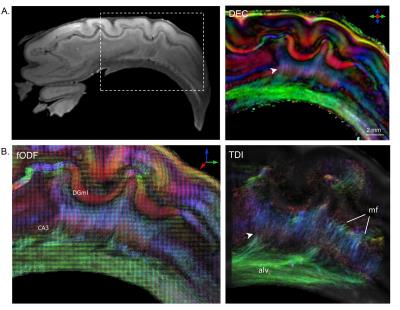
Fig. 3 depicts the level of intra-hippocampal subfield connectivity that could be resolved with HARDI. The DEC map (Fig. 3A) reveals very fine microscopic fibers projecting between the DGgr and the CA3 subfields (white arrowhead). Granule cell unmyelinated axons, called mossy fibers, project to the CA3 pyramidal neurons and form a part of the hippocampal trisynaptic circuit8. These microscopic mossy fibers could be distinctly delineated in the DEC map (arrowhead in Fig. 3A) and reconstructed using probabilistic tractography (TDI map in Fig. 3B). Further, axonal fibers in the trisynaptic circuit extend from CA3 to the alveus. These microscopic projections from CA3 to alvear white matter could also be reconstructed using probabilistic tractography, and are shown in the TDI map (Fig. 3B).
Conclusion
The results of this work demonstrate intricate microstructural organization of the human hippocampus delineated with high angular and spatial resolution dMRI, which provided unique insights into its complex 3D structure and subfield connectivity. These results have important future applications to allow 3D mapping of hippocampal atrophy and mossy fiber disruption in Alzheimer’s disease and other neurological disorders.Acknowledgements
This work was supported by National Institute of Health (NIH) grants R03EB017806 and R21NS096249References
1. Duvernoy HM, Cattin F, Risold PY. The Human Hippocampus: Functional Anatomy, Vascularization and Serial Sections with MRI: Springer Berlin Heidelberg; 2013.
2. Augustinack JC, Magnain C, Reuter M, van der Kouwe AJ, Boas D, Fischl B. MRI parcellation of ex vivo medial temporal lobe. NeuroImage 2014;93 Pt 2:252-259.
3. Yushkevich PA, Avants BB, Pluta J, Das S, Minkoff D, Mechanic-Hamilton D, Glynn S, Pickup S, Liu W, Gee JC, Grossman M, Detre JA. A high-resolution computational atlas of the human hippocampus from postmortem magnetic resonance imaging at 9.4 T. NeuroImage 2009;44(2):385-398.
4. Iglesias JE, Augustinack JC, Nguyen K, Player CM, Player A, Wright M, Roy N, Frosch MP, McKee AC, Wald LL, Fischl B, Van Leemput K. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. NeuroImage 2015;115:117-137.
5. Tournier JD, Calamante F, Connelly A. Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. NeuroImage 2007;35(4):1459-1472.
6. Calamante F, Tournier JD, Jackson GD, Connelly A. Track-density imaging (TDI): super-resolution white matter imaging using whole-brain track-density mapping. NeuroImage 2010;53(4):1233-1243.
7. Behrens TE, Woolrich MW, Jenkinson M, Johansen-Berg H, Nunes RG, Clare S, Matthews PM, Brady JM, Smith SM. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn Reson Med 2003;50(5):1077-1088.
8. Amaral DG, Insausti R. The hippocampal formation. In: Paxinos G, editor. The human nervous system: Academic Press, San Diego, CA.; 1990.
Figures